Description
Green fluorescent protein (GFP) is one of the most common fluorophores used to label cells and proteins in C. elegans. Previous artificial protein evolution strategy has generated a variant of eGFP, GFPnovo2, which is 3.3 times brighter than the original eGFP in the DT40 cell line (Arakawa et al, 2008). GFPnovo2 carries four mutations from the original eGFP (Y145F, V163A, S202T, L221V), and has the same excitation/emission wavelengths as the original eGFP.
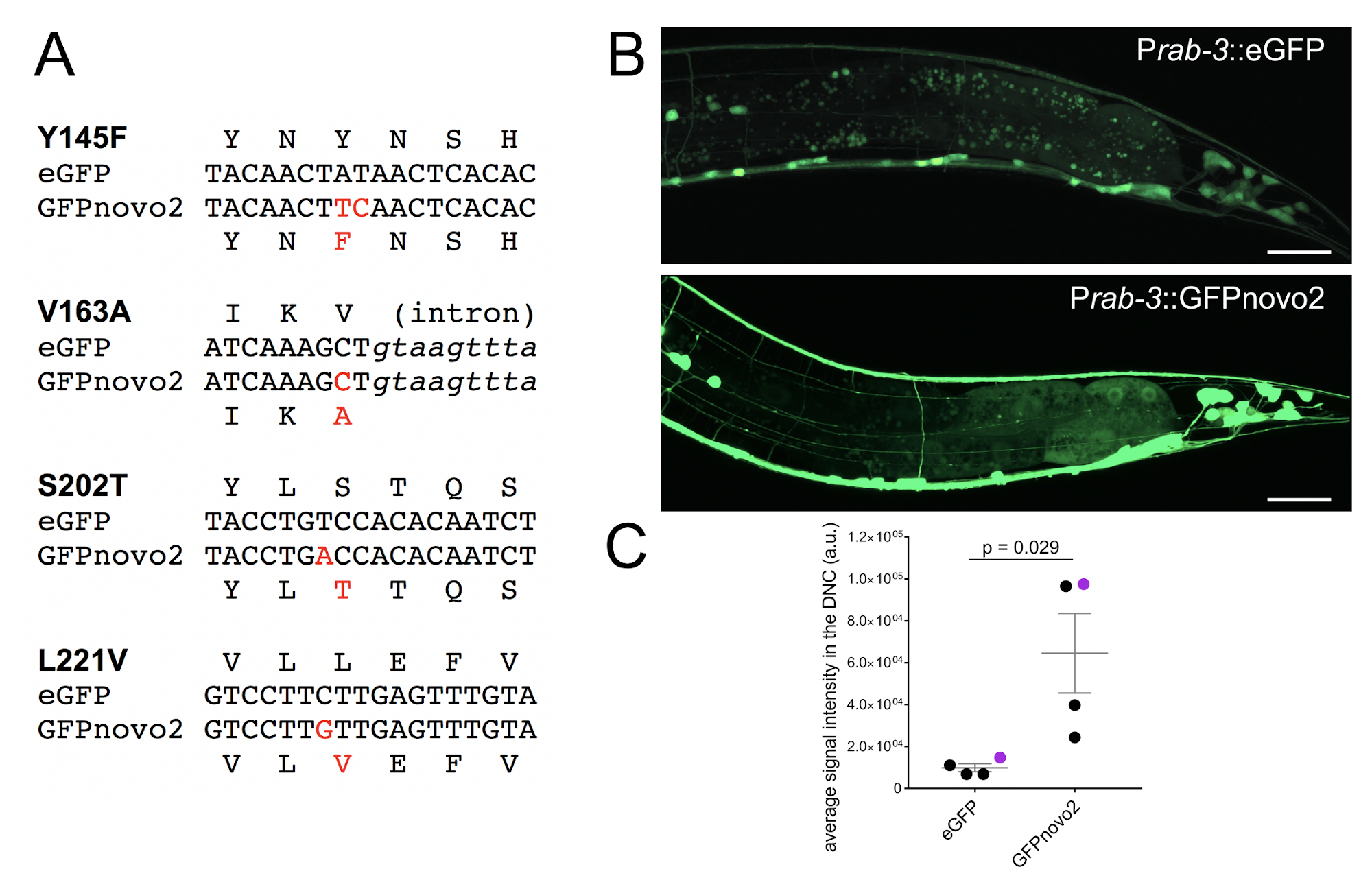
Here we compared the brightness of GFPnovo2, which was generated by introducing above mentioned mutations in the pSM (eGFP_unc-54 3’ utr: kind gift from Cori Bargmann) vector (Figure 1A), with that of original eGFP in the C. elegans nervous system. Under the fluorescent dissection scope, all four GFPnovo2 lines with high transmission rate (over 80%) we examined had brighter fluorescent signal at all developmental stages (late embryo to adult) than five eGFP lines with the similar high transmission rate. As axons and dendrites are thin, it is often difficult to visualize when the copy number of the transgene is low. Indeed, we observed fewer neurites when we labeled neurons with the low dose of eGFP (1ng/ml) expressed under the pan-neuronal promoter, Prab-3 (Figure 1B, top). In contrast, GFPnovo2 was considerably brighter than eGFP and nicely labeled neurites when injected at the same concentration (1ng/ml) (Figure 1B, bottom). As a result, the average signal intensities of the dorsal nerve cord, which contains only neurites, was significantly and consistently brighter in the animals expressing GFPnovo2 than those expressing eGFP (Figure 1C). The variation among the animals expressing GFPnovo2 is likely due to the mosaic nature of the extra-chromosomal array. Nevertheless, all GFPnovo2 animals had a brighter signal than eGFP animals. Similarly, we were able to label the entire axon of DA9 neuron with GFPnovo2 expressed under the DA9 specific promoter, itr-1 (Chen et al., 2018), including the axonal tip which was not easy to detect with eGFP (unpublished). We did not notice detectable photobleaching while examining animals expressing GFPnovo2 under the fluorescent compound microscope or the confocal microscope, suggesting that GFPnovo2 is at least as stable as eGFP.
Qualifiers
- In this manuscript, we did not conduct definitive comparison between GFPnovo2 and eGFP using single-copy integration of the transgene. The comparison is therefore rather subjective, even though the difference is very noticeable at the dissection microscopy level.
- We have not tested whether the reagents for GFP such as antibodies and GFP nanobody::ZIF-1 for tissue specific protein degradation (Wang et al., 2017) could be used with GFPnovo2. Arakawa et al. suggested that the overall structures of GFPnovo2 is unlikely to be changed much.
- We have so far tested GFPnovo2 only in the nervous system (the leaky expression of the Prab-3 and Pitr-1 suggests GFPnovo2 is also brighter in the hindgut). It is therefore not known if GFPnovo2 is brighter in all tissues at any given developmental stage.
Conclusion
GFPnovo2 could be the useful option for labeling cells and proteins in C. elegans, especially when the copy number of the transgene is low such as single-copy integration or CRISPR-mediated tagging of the endogenous proteins.
Reagents
Plasmid: pSM.GFPnovo2 Addgene #116943
Strains: UJ1000: mizEx328 [Prab-3::eGFP;Podr-1::GFP]; UJ1001: mizEx329 [Prab-3::GFPnovo2;Podr-1::GFP].
References
Funding
This work is funded by NSERC (RGPIN-2015-04022)
Reviewed By
Jordan WardHistory
Received: August 28, 2018Accepted: September 6, 2018
Published: September 6, 2018
Copyright
© 2018 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Hendi, A; Mizumoto, K (2018). GFPnovo2, a brighter GFP variant for in vivo labeling in C. elegans. microPublication Biology. 10.17912/49YB-7K39.Download: RIS BibTeX