Molecular and Cellular Physiology, Stanford University, Stanford, CA
Description
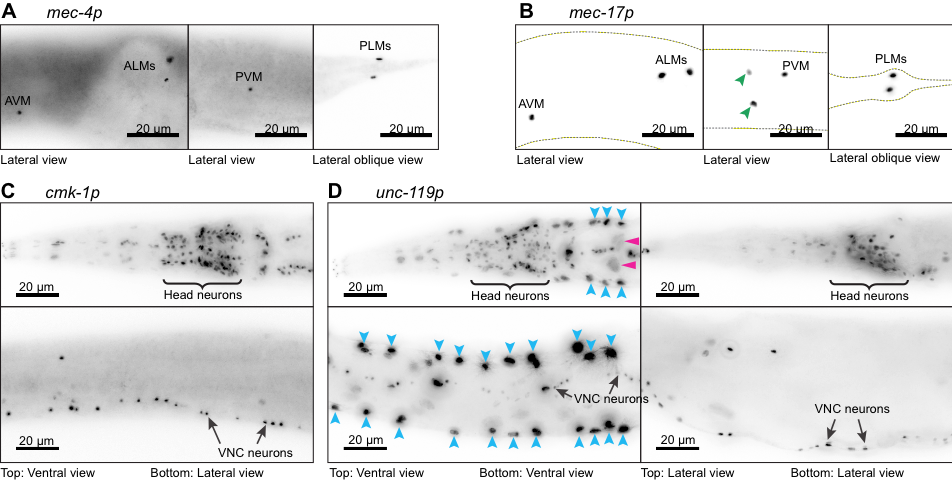
Four transgenes designed to drive expression of Cre recombinase and a nuclear-localized blue fluorescent protein (<Neuron-specific promoter>::Cre::F2A::BFP::AID::NLS::tbb-2 3’UTR) were expressed under the control of neuron-specific promoters. All transgenes were inserted into the ttTi5605 transposon insertion site in chromosome II of wild-type C. elegans animals (N2) using CRISPR/Cas9-mediated genome editing, equalizing any potential positional effects. We observed very faint BFP signal in live worms from the signal copy transgene insertions. Additionally, the signal was further masked by bright autofluorescence from the intestine. To observe the cells expressing these transgenes we performed immunofluorescence experiments using primary antibody against BFP. Using this assay, we studied the expression from touch receptor neuron (TRN)-specific promoters, mec-4p (A) and mec-17p (B) and pan-neuronal promoters, cmk-1p (C) and unc-119p (D). As expected, BFP expression driven by the mec-4promoter is observed exclusively in the six TRN nuclei (A). However, under mec-17 promoter, BFP expression is observed in two additional neuronal nuclei (B, green arrowheads) besides the six TRN nuclei (B). These two neurons are likely to be PVD or PDE neurons based upon the location of their nuclei relative to the PVM nucleus. In case of the pan-neuronal promoters, expression under cmk-1 and unc-119 promoters is observed in most, if not all neuronal nuclei, including in many head neurons (C and D top) and ventral nerve cord (VNC) neurons (C and D bottom). However, in a subpopulation of animals expressing BFP under the unc-119 promoter, intense fluorescence was observed in several additional larger nuclei along the lateral sides (D, blue arrowheads), which we identify as hypodermal cells and faint signals in even larger nuclei (D, pink arrowheads), which we identify as intestinal cells. We did not observe any fluorescence in our control animals (N2) which were treated similarly (data not shown). While this experiment confirmed expected expression patterns for mec-4p and cmk-1p, additional expression in other neurons for mec-17p and other tissue types in unc-119p promoters could reflect the specific promoter sequences used in this study or that low levels of expression in these unexpected cells were not readily detected from direct observation of fluorescent protein expression analyzed previously.
Methods
Request a detailed protocolAll worm strains were grown at 20 °C on NGM plates containing OP50-1. Well-fed adult hermaphrodites were fixed according to the protocol described by Michael Koelle (1), with the following modifications:
- All steps after the initial washing off worms from the NGM plates were performed in 48-well plates instead of microcentrifuge tubes.
- Instead of centrifugation, worms were allowed to settle by gravity to reduce shear-induced rupturing of worms.
- Primary antibody against BFP (rabbit polyclonal antibody, Evrogen AB233) was used at 1:5000 dilution.
- F(ab’)2-Goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 568 conjugate (Invitrogen, A-21069), was used at 1:4000 dilution.
Reagents
The following strains are used in this study.
GN706 (pgSi101[mec-4p::Cre::F2A::BFP::AID::NLS::tbb-2 3’UTR]) II
GN707 (pgSi102[mec-17p::Cre::F2A::BFP::AID::NLS::tbb-2 3’UTR]) II
GN751 (pgSi114[cmk-1p::Cre::F2A::BFP::AID::NLS::tbb-2 3’UTR]) II
GN730 (pgSi110[Cbr-unc-119p::Cre::F2A::BFP::AID::NLS::tbb-2 3’UTR]) II
The following promoter sequences was used in this study.
mec-4p: 1020 bp region immediately upstream of mec-4 start codon (2)
mec-17p: 868 bp region immediately upstream of mec-17 start codon
cmk-1p: 2000 bp region immediately upstream of cmk-1 start codon (3)
Cbr-unc-119p: 988 bp region immediately upstream of Cbr-unc-119 start codon (4)
Full sequences of all reported alleles are available upon request.
References
Funding
Work funded by grants from NIH (R01NS092099, R01NS105092 to MBG) and a 2017 Amgen Scholarship (SL).
Reviewed By
AnonymousHistory
Received: May 31, 2018Accepted: July 18, 2018
Published: July 20, 2018
Copyright
© 2018 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Lear, SK; Das, A; Goodman, MB (2018). Immunofluorescence reveals neuron-specific promoter activity in non-neuronal cells. microPublication Biology. 10.17912/8FDA-CK77.Download: RIS BibTeX