Howard Hughes Medical Institute
Description
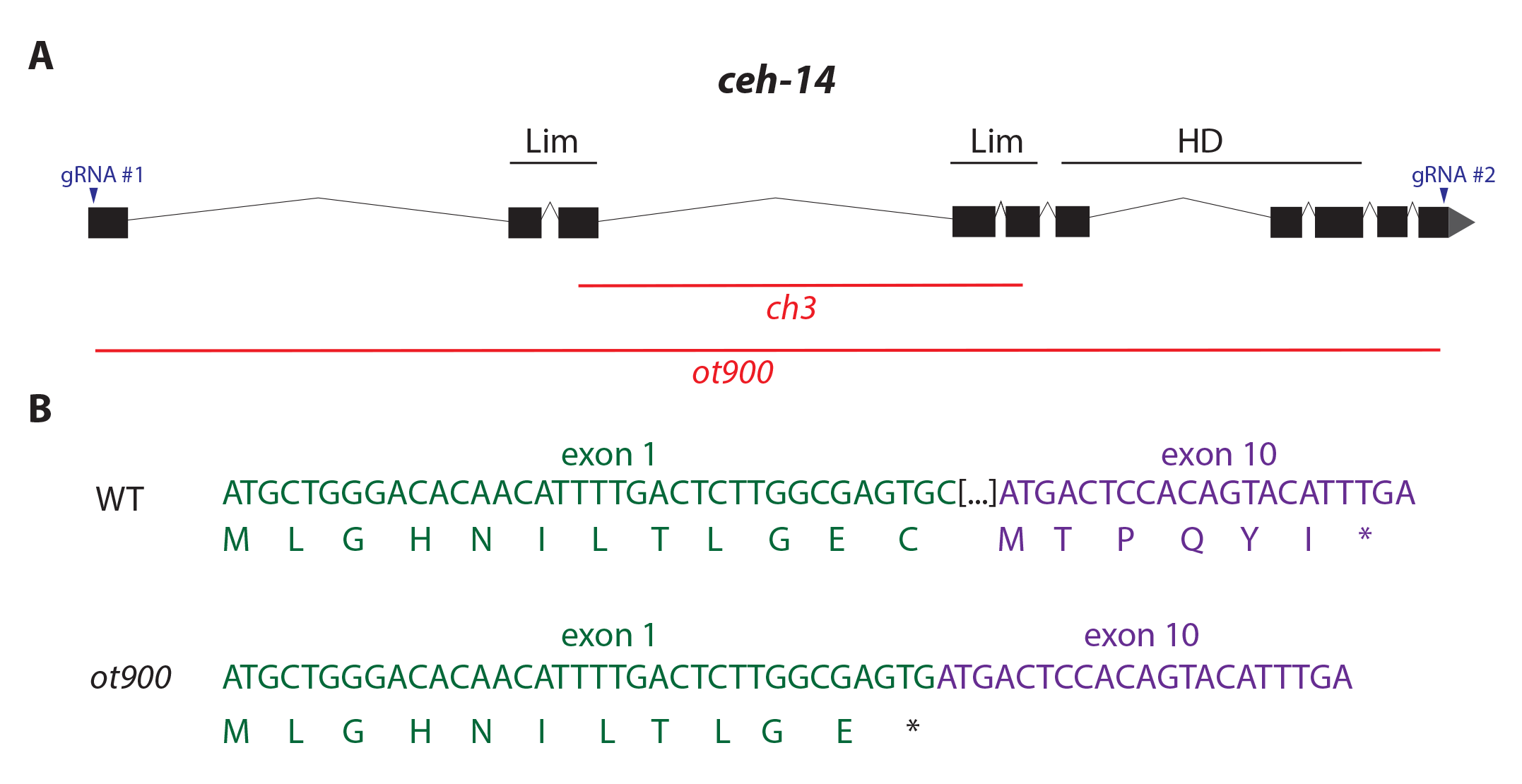
We have generated a novel null allele, ot900, of the C. elegans LIM homeodomain gene ceh-14. All existing deletion alleles of ceh-14 were generated using a Tc1 transposon insertion, one of which, ch3, is a putative null based on the resulting frame-shift and a reduction in detectable mRNA and protein levels (Cassata et al. 2000). However, in our hands we have noticed a propensity of the ch3 allele to revert to wild-type after several generations, perhaps due to a more complex rearrangement of the locus caused by the Tc1 transposon than previously appreciated.
To avoid potential complexities of working with the ch3 allele, we used CRISPR/Cas9 to generate a new null allele of ceh-14. The ot900 allele was isolated with gRNAs targeted to the first and last exons of ceh-14 (gRNA #1: CTTGGCGAGTGCGATGAGC, gRNA #2: GTACTGTGGAGTCATGTGT; see Fig. 1), and then PCR screening for resulting large deletion alleles. Our primers for PCR genotyping of the deletion are 5’-CTCAACTAAATCGTCAGAATCGTC-3’ and 5’-GAGATGTATGAAACGAGCGAGCG-3’ which generate a 620bp product in the context of the ot900 deletion. The ceh-14 locus in the ot900 allele is only capable of generating an 11aa peptide due to the remaining bases in the first exon, before reaching a premature stop codon (Fig 1B).
Previous studies have implicated ceh-14 in differentiation and function of the phasmid neurons, but have shown incomplete penetrance and variability in phenotypes resulting from the ceh-14 (ch3) allele (Kagoshima et al. 2013, Serrano-Saiz et al. 2015). We used the srg-13 marker of PHA identity (which was expressed in 20/20 wild-type animals) to assess whether phasmid neuron differentiation is more severely affected in ot900 than ch3, and found that this marker was indeed lost in 100% of animals (expressed in 0/15 animals), despite not finding an appreciable effect in ceh-14 (ch3) (expressed in 15/15 animals). This suggests that some of the variability in phenotype may be the result of the ch3 allele itself, rather than a property of CEH-14 function.
Reagents
OH15422 ceh-14 (ot900) X. Will be available at CGC.
References
Funding
This work was supported by the HHMI and NIH (R37NS039996, O.H., F31NS096863, E.A.B.)
Reviewed By
Thomas BürglinHistory
Received: October 10, 2018Accepted: October 18, 2018
Published: October 18, 2018
Copyright
© 2018 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Bayer, E; Hobert, O (2018). A novel null allele of C. elegans gene ceh-14. microPublication Biology. 10.17912/g434-3d85.Download: RIS BibTeX