Département de Biologie Médicale, Université du Québec à Trois-Rivières, Trois-Rivières, Canada.
Département de Pathologie et Biologie Cellulaire, Institut de Recherche en Immunologie et en Cancérologie (IRIC), Université de Montréal, Montréal, Canada.
Description
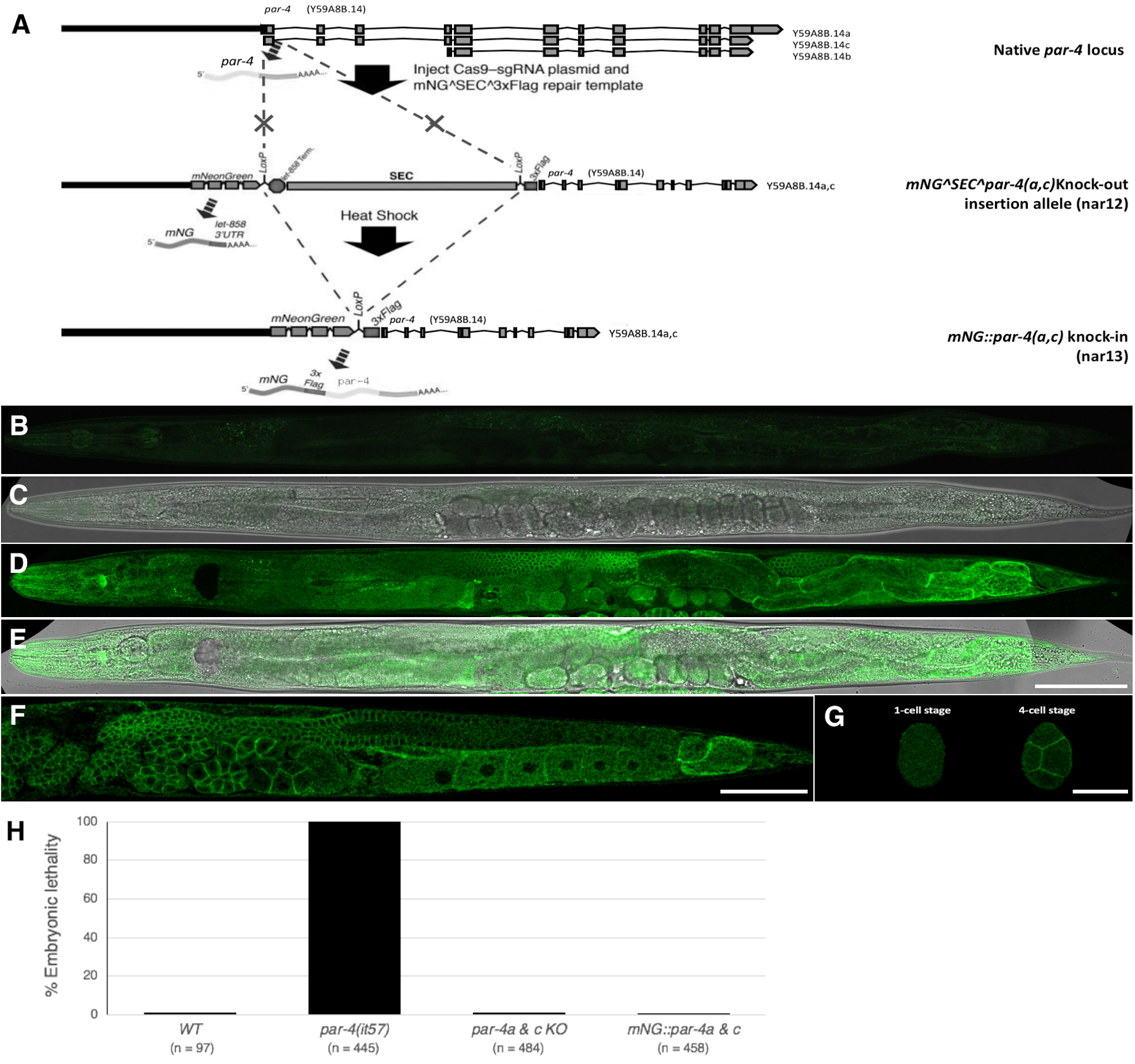
PAR-4/LKB1 is maternally expressed and required for the asymmetrical distribution of early embryonic determinants and viability in C. elegans (Morton et al. 1992; Watts et al. 2000; Tenlen et al. 2008). It is also implicated in a variety of postembryonic processes, including germline stem cell quiescence (Narbonne and Roy 2006; Narbonne et al. 2017), neuronal growth and polarity (Kim et al. 2010; Teichmann and Shen 2011), cytoskeletal rearrangements (Narbonne et al. 2010; Chartier et al. 2011), and metabolism (Narbonne and Roy 2009). Its expression pattern and subcellular localization has been determined in fixed animals by antibody staining (Watts et al. 2000). Here, we used CRISPR/Cas9-mediated genome editing to fluorescently label the two longest endogenous PAR-4 isoforms, PAR-4A and PAR-4C, with monomeric Neon Green (mNG) (Shaner et al. 2013), using the self-excising cassette (SEC) system (Dickinson et al. 2015) (Fig. 1A). We found ubiquitous mNG::par-4a & c expression and cytoplasmic and cortical enrichment of mNG::PAR-4A & C proteins in the germ line and early embryos (Fig. 1B-H), as previously described (Watts et al. 2000). Interestingly, we find that mNG::PAR-4A & C cortical enrichment is transiently lost in the pachytene area of the germ line (Fig. 1F), although it remains unclear whether this is functionally relevant.
The intermediate strain that still contains the SEC (Dickinson et al. 2015) is predicted to be null for par-4a & c, potentially leaving the shorter par-4b isoform functional. To evaluate the requirement for par-4a & c we examined the embryonic lethality (at 25°C) of the generated SEC-containing and SEC-excised strains. We found that the self-progeny of homozygous SEC-containing (e.g. par-4a & c null) animals is viable (Fig. 1H). This suggests that par-4b alone is sufficient to establish embryonic polarity and sustain the essential function of par-4. Consistent with this, to our knowledge, all existing par-4 alleles that impair embryonic development disrupt par-4b (Morton et al. 1992; Watts et al. 2000).
Reagents
Nematodes were cultured on standard NGM plates with E. coli (OP50) and maintained at 15 °C unless otherwise specified. N2 (Bristol) was used as wild-type. The following strains were also used: KK300: par-4(it57)V, UTR43: par-4(nar12[Ppar-4a::mNG + loxP sqt-1(gf) Hygromycin (+) loxP 3X FLAG])V, UTR45: par-4(nar13[mNG :: par-4a & c])V. UTR43 and UTR45 will be made available through the CGC.
pDD162 was modified as described (Paix et al. 2014) to generate two sgRNAs using the following primers (all 5’->3’): fwd Q5 1 gtgctcccgaggatgtcgagttttagagctagaaatagcaagt and fwd Q5 2 atgctccgtcgacatcctcgttttagagctagaaatagcaagt. pDD268 was modified as described (Dickinson et al. 2015) to generate the N-terminal mNG::SEC repair template using the following primers: 5’ arm par-4 fwd acgttgtaaaacgacggccagtcgccggcaatttggtcgtttttggggtt, 5’ arm par-4 rev catgttgtcctcctctcccttggagaccattgaagagagctctgaaattttt, 3’ arm par-4 fwd cgtgattacaaggatgacgatgacaagagaatggacgcaccgtcaacttcatcaggagcacaaagcaaacttctg, and 3’ arm par-4 rev tcacacaggaaacagctatgaccatgttatttccgaaaattgaacgattttt. Modified vector DNA sequences were confirmed by Sanger sequencing. The two sgRNA guide vectors were microinjected together with the repair template in wild-type animals that had been subjected to cku-80(RNAi) (Ward 2015) and a single par-4 CRISPRed line was obtained; the SEC was excised as described (Dickinson et al. 2015).
All images were acquired from paralyzed live animals (0.1% W/V tetramizole in M9) with a Leica SP8 confocal microscope using the same parameters and were processed identically. Whole animals were stitched (Preibisch et al. 2009) and straightened using ImageJ.
Acknowledgments
We thank Dan Dickinson and Bob Goldstein for providing pDD268 and the CGC (funded by NIH Office of Infrastructure Programs P40 OD010440) for strains.
References
Funding
This work was funded by CIHR grant PJT-153283 to JCL, and FRQ-NT (NC-205752) and CFI (36916) grants to PN. PN is a Junior 1 FRQ-S Research Scholar.
Reviewed By
AnonymousHistory
Received: October 11, 2018Accepted: November 12, 2018
Published: November 13, 2018
Copyright
© 2018 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Roy, V; Gagné, O; Hamiche, K; Labbé, JC; Narbonne, P (2018). Expression pattern of endogenous PAR-4A & C after CRISPR/Cas9-mediated genome editing. microPublication Biology. 10.17912/micropub.biology.000075.Download: RIS BibTeX