Division of Gerontology and Geriatric Medicine, Department of Medicine, University of Washington, Seattle, WA 98104, USA
Description
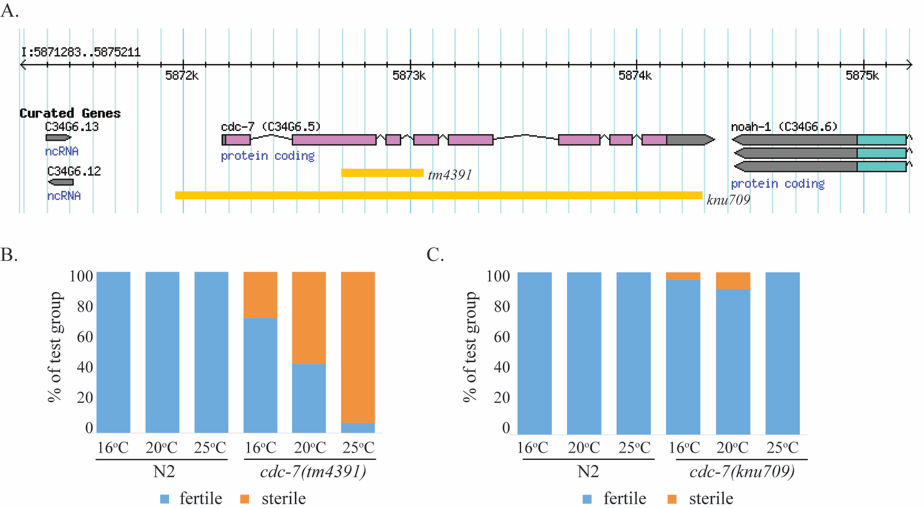
CDC7 regulates both DNA replication initiation and checkpoint-regulated progression of the cell cycle during the G1/S phase, contributes to DNA recombination and damage repair, and is an essential gene in many species (Yamada et al. 2014). In C. elegans, there has been a single characterized deletion allele available, tm4391, impacting the 8-exon coding sequence of cdc-7. tm4391 is missing 315 bp including part of exon 2, all of exon 3, and part of exon 4, and resulting in a downstream frame shift. cdc-7(tm4391) worms are temperature sensitive sterile, with worse fertility at higher temperatures (Figure 1B). We generated a novel deletion allele knu709, which is an approximately 2200 bp deletion spanning 150 bp upstream of exon 1 to about 150 bp after exon 8 and removing the entire cdc-7coding sequence. We tested knu709 for temperature sensitive sterility. Surprisingly, this strain is not temperature sensitive sterile (Figure 1C), indicating that the fertility defects of tm4391 may be due to either a closely linked mutation in another gene or an unexpected gain-of-function or partial loss-of-function phenotype of cdc-7(tm4391) not recapitulated by a true loss-of-function mutation.
Reagents
Strains:
CK609 cdc-7(tm4391) I – outcrossed 4x to N2.
NLS1 cdc-7(knu709) I – outcrossed 3x to N2. Strain will be sent to CGC.
Methods
Request a detailed protocolGenotyping:
Oligos used for genotyping tm4391: Mix all 3 primers in a single reaction. Expect sizes 757bp and 270bp WT, 400bp mutant.
NL93 (F): tcagtgcaacatgcagaaca
NL94 (R): tgacacaaaccaatcccaaa
NL187 (internal deletion): ATGCGACAGCATAAAGCAAA
Oligos used for genotyping knu709: Mix all 3 primers in a single reaction. Expect sizes 2848bp and 823bp WT, 565bp mutant.
NL429 (F): cccgtatcacacactcatcg
NL430 (R): attgctaaaacccgcagaaa
NL431 (internal deletion): ggaaacgtaccctcgcctat
Fertility Assays
Synchronized populations of C. elegans were established by treating with alkaline hypochlorite solution (bleaching) (Porta-de-la-Riva et al. 2012). Eggs were grown at 16oC to L4 stage at which point worms were singled onto 60mm NGM plates seeded with OP50. Plates with singled worms were then divided into 3 groups of approximately 50 per strain. Groups were transferred to the assay temperature: either 25oC, 20oC or 16oC. After 1 week, plates were scored for the presence of progeny. The absence of progeny was scored as sterile.
CRISPR/Cas9
COP1803 cdc-7(knu709) was generated by Knudra Transgenics/ NemaMetrix (Eugene, OR), and the deletion end-points confirmed by sequencing. 3-frame stop insertion sequence at breakpoints of deletion: TAAATAAATAAACTCGAG.
Acknowledgments
We thank Drs. Brian Kraemer and Tom Bird for expert advice. We thank the National Bioresource Project (Japan) for providing strains. We thank WormBase for C. elegans model organism information. We thank the Department of Veterans Affairs for funding.
Author Contributions: N.L. was involved in this project conceptualization, formal analysis, methodology, project administration, resources, supervision, visualization and writing. H.C. was involved in project conceptualization, formal analysis, investigation, methodology, project administration, visualization and writing.
References
Funding
Department of Veterans Affairs Career Development Award 2 #I01BX007080 and Merit Review Grant #I01BX004044 to N.L.
Reviewed By
Kelly LiuHistory
Received: December 28, 2018Accepted: January 3, 2019
Published: January 3, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Currey, H; Liachko, N (2019). A CRISPR/Cas9-generated cdc-7 loss of function mutation does not cause temperature-dependent fertility defects. microPublication Biology. 10.17912/micropub.biology.000085.Download: RIS BibTeX