Harvard Medical School, Boston, MA, 02115
Description
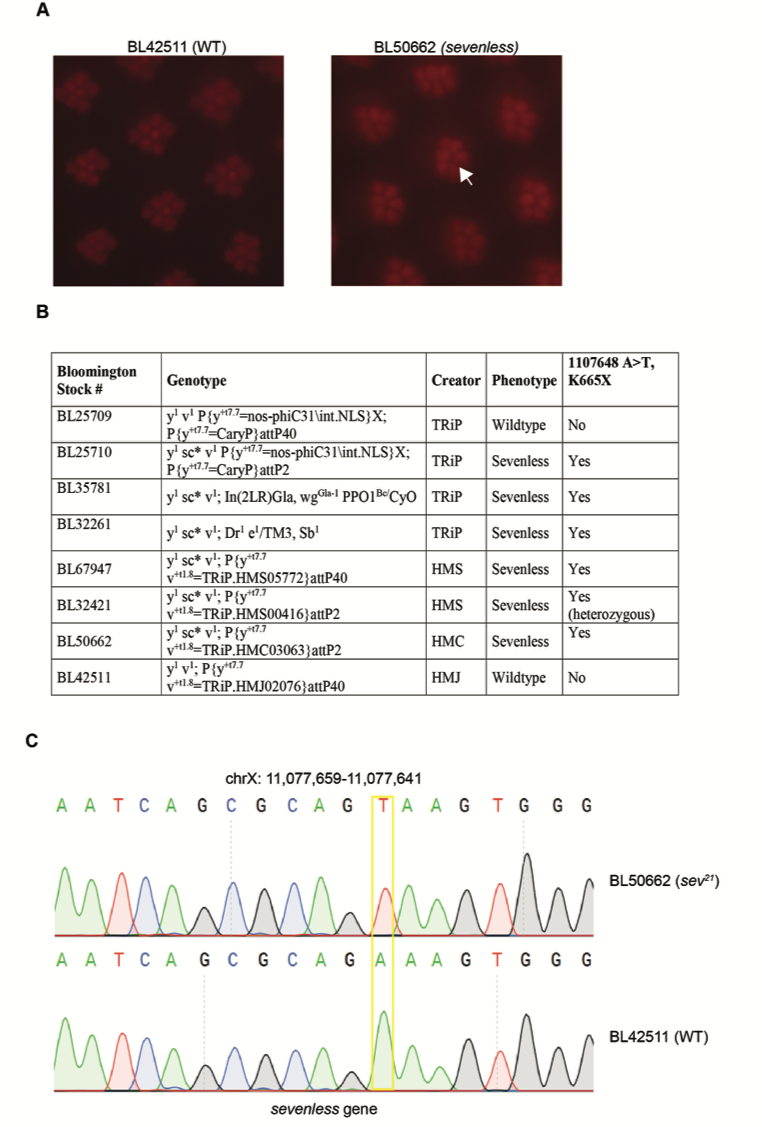
The Transgenic RNAi Project (TRiP) has generated more than 12,000 transgenic RNAi fly stocks that have been distributed to the community via the Bloomington Drosophila Stock Center (Ni et al. 2007; Ni et al. 2011; Perkins et al. 2015). These stocks express long double-stranded RNA hairpins (dsRNAs) or short RNA hairpins (shRNAs) under GAL4/UAS control (Brand and Perrimon 1993), and provide powerful tools for targeted genetic screens. Unexpectedly, as part of a genetic screen examining retinal degeneration in flies, we identified a defect in eye development associated with many of the TRiP stocks. Drosophila have a compound eye composed of repeating units, termed ommatidia, that each contain eight photoreceptor cells (R cells 1 – 8) (Ready, Hansen and Benzer 1976). The light-sensing organelle, the rhabdomere, in seven of these photoreceptors can be directly visualized in wild-type flies using light microscopy either by optical neutralization or by examining the deep pseudopupil; R7/R8 are stacked on top of each other so only one is visible in a given vertical plane (Franceschini and Kirschfeld 1971).Whereas seven rhabdomeres could be counted per ommatidium in wild-type flies (Fig. 1A), a subset of the TRiP lines tested show characteristic loss of a single rhabdomere (Fig. 1A, Fig. 1B). This single photoreceptor loss phenotype is reminiscent of sevenless (sev) mutants; sev (FBgn0003366) encodes a receptor tyrosine kinase essential for development of R7; thus, loss of function sev mutations result in ommatidia that lack R7 (Harris et al. 1976; Simon, Bowtell and Rubin 1989). Preliminary observations suggested that the sev phenotype was X-linked and observed only in TRiP stocks containing a scute (sc) allele of unknown origin denoted sc*. Whole genome sequencing data for one of the TRiP stocks with the X chromosome containing this sc allele (y1 sc* v1) revealed the presence of an A>T mutation at position X:1107648 in sev, which would result in a premature stop codon at K665X. We tested several of the TRiP stocks that showed the sev phenotype using PCR sequencing, and found that all contained the same mutation (Fig. 1C). We have named this new allele sev21. We note that we observed both sev21 and wild-type flies in BL32421, suggesting that this stock is mixed. Since this premature stop codon would result in a severely truncated protein, it is likely that the sev21 allele would represent a loss-of-function mutation. Supporting this, our newly identified sev21 allele did not complement the known sev14 loss-of-function allele (BL67947, BL32421, BL50662). Since stocks generated by the TRiP at Harvard Medical School and their collaborators at Tsinghua University show the sev phenotype, but stocks generated by TRiP collaborators at the National Institute of Genetics in Japan do not, we suspected that the mutation was likely present in the stocks used to balance the TRiP lines (BL35781 and BL32261). PCR sequencing revealed that both of these stocks carry the sev21 allele. Together, these data show that many of the TRiP RNAi stocks balanced with BL35781 or BL32261 contain a newly identified loss-of-function sev allele, sev21. TRiP stocks containing this sev21 allele, including both RNAi and sgRNA lines (TRiP-CRISPR Overexpression and KnockOut) (Port et al. 2014; Jia et al. 2018), will be annotated on Flybase and at BDSC. The presence of the sev21 mutation will not generally affect the use of these stocks, as the X chromosome is typically segregated out or heterozygous during experiments.
Reagents
BL25709 (RRID:BDSC_25709): y1 v1 P{y+t7.7=nos-phiC31\int.NLS}X; P{y+t7.7=CaryP}attP40
BL25710 (RRID:BDSC_25710): y1 sc* v1 P{y+t7.7=nos-phiC31\int.NLS}X; P{y+t7.7=CaryP}attP2
BL35781 (RRID:BDSC_35781): y1 sc* v1; In(2LR)Gla, wgGla-1 PPO1Bc/CyO
BL32261 (RRID:BDSC_32261): y1 sc* v1; Dr1 e1/TM3, Sb1
BL67947 (RRID:BDSC_67947): y1 sc* v1; P{y+t7.7 v+t1.8=TRiP.HMS05772}attP40
BL32421 (RRID:BDSC_32421): y1 sc* v1; P{ y+t7.7 v+t1.8=TRiP.HMS00416}attP2
BL50662 (RRID:BDSC_50662): y1 sc* v1; P{ y+t7.7 v+t1.8=TRiP.HMC03063}attP2
BL42511 (RRID:BDSC_42511): y1 v1; P{ y+t7.7 v+t1.8=TRiP.HMJ02076}attP40
BL5690 (RRID:BDSC_5690): sev14; Ras85De2F/TM3, Sb1
BL5691 (RRID:BDSC_5691): sev14; drke0A/CyO
Acknowledgments
We would like to thank Dr. Andrew Zelhof for confirming the presence of the sev phenotype, Dr. Yanui Hu for compiling list of variants in sev from TRiP stock whole genome sequence, and Dr. Kevin Cook and Dr. Annette Parks for providing Drosophila stocks and for their input on the manuscript.
References
Funding
Research reported in this publication was supported by the National Eye Institute of the NIH under Award Number R01EY024905 to VW.
Reviewed By
AnonymousHistory
Received: March 19, 2019Accepted: March 24, 2019
Published: April 4, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Escobedo, SE; Zirin, J; Weake, VM (2019). TRiP stocks contain a previously uncharacterized loss-of-function sevenless allele. microPublication Biology. 10.17912/micropub.biology.000097.Download: RIS BibTeX