Department of Biochemistry and Molecular Biophysics, Department of Biological Sciences, Howard Hughes Medical Institute, Columbia University, New York, USA
Description
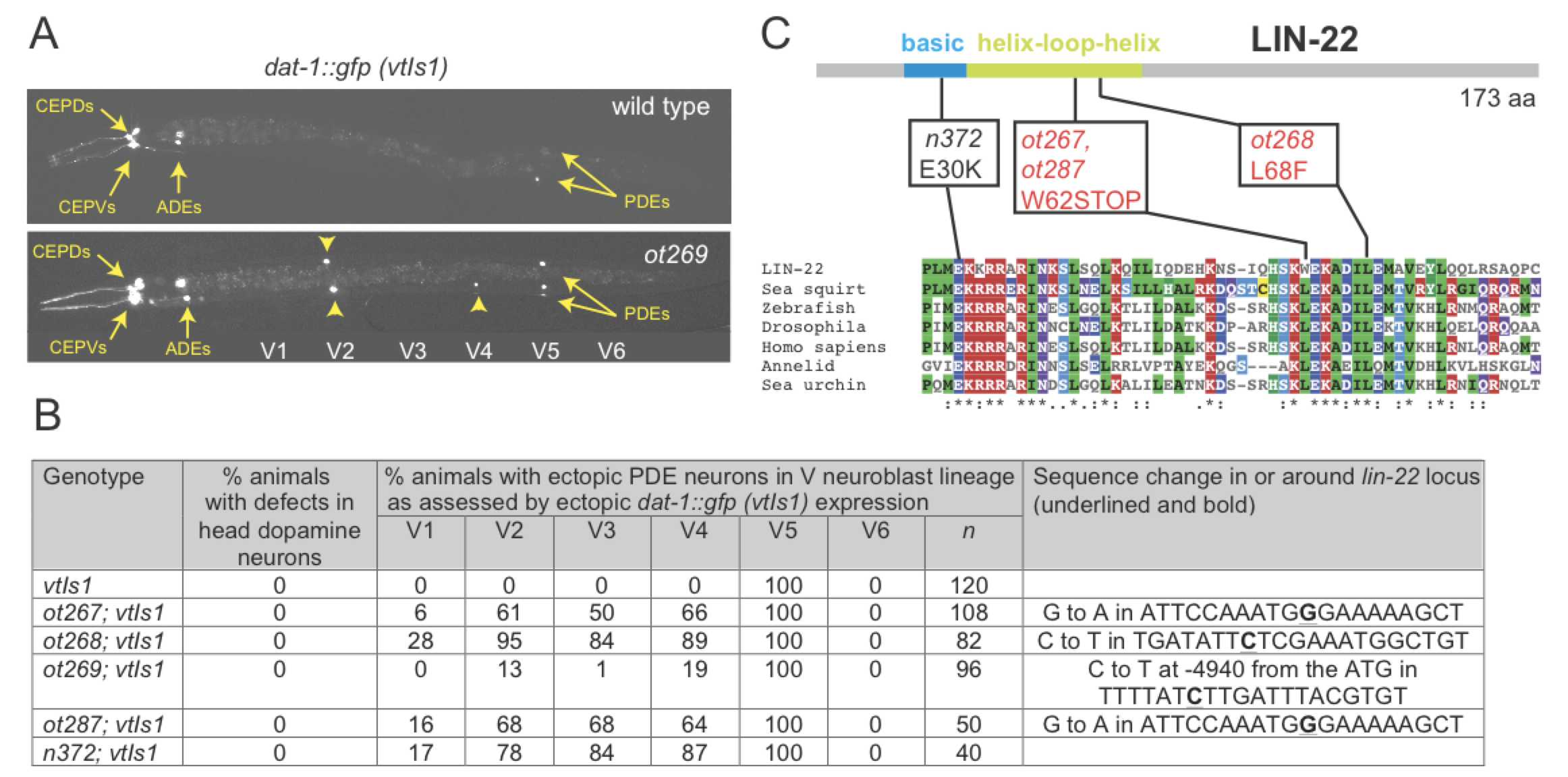
We screened for mutants that affect expression of dopaminergic neuron identity, using a transcriptional reporter for expression of the dopamine transporter dat-1. We previously published and characterized a number of mutants that affect dat-1 expression in different neuron types (Doitsidou et al., 2008). Four alleles that we did not publish in our original screening paper are described here. While wild-type animals only display a single dat-1::gfp(+) neuron pair in the midbody region, the PDE neuron pair from the postdeirid lineage, all 4 mutant alleles display ectopic dat-1::gfp expression along the anterior/posterior axis of the animal (Fig.1A,B). Postdeirid lineage duplication defects were previously described in animals lacking the bHLH transcription factor lin-22/Hairy (Wrischnik and Kenyon, 1997). We find that the canonical lin-22 allele, n372, indeed displays dat-1::gfp expression defects similar to those observed in our mutants (Fig.1B). We sequenced the lin-22 locus in all of our four, independently isolated alleles. Two of them are premature stop codons, one is a missense mutation affecting a conserved leucine residue and all display a similar penetrance of defects (Fig.1B,C). The fourth and weakest allele, ot269, displayed no sequence alteration in the lin-22 coding sequence or in exon/intron boundaries. ot269 failed to complement ot267, ot268, ot287 and the canonical lin-22 allele n372. Furthermore, the ot269 phenotype was rescued by injection of the fosmid WRM0627dG07, which contains lin-22 and one additional complete gene. We found that ot269 harbors a single nucleotide change in the upstream intergenic region of lin-22, almost 5kb away from the start of the gene (sequence change shown in Fig.1B). Subsequent work has shown that this mutation affects a binding site for a GATA transcription factor (Katsanos et al. 2017).
Reagents
OH4265 lin-22(ot267);vtIs1
OH4270 lin-22(ot268);vtIs1
OH4271 lin-22(ot269);vtIs1
OH4320 lin-22(ot287);vtIs1
Strains are available at the CGC.
Acknowledgments
We thank Chi Chen for injections, Andrea J. Pretorian and Albert Lee for help with screening, Alexander Boyanov and Berta Vidal for help with sequencing.
References
Funding
This work was funded was funded by the US National Institutes of Health (R01NS050266), the Howard Hughes Medical Institute and a European Molecular Biology Organization long-term fellowship to M.D.
Reviewed By
Jintao LuoHistory
Received: April 4, 2019Accepted: April 19, 2019
Published: April 19, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Doitsidou, M; Hobert, O (2019). New alleles of the lin-22/Hairy bHLH transcription factor. microPublication Biology. 10.17912/micropub.biology.000111.Download: RIS BibTeX