Description
The genes encoding transcription factors that initiate the terminal differentiation programs within individual neurons have been termed “terminal selectors” (Hobert 2008). They are considered master regulators of neuronal identity because they integrate upstream lineage inputs to ultimately drive (directly and/or indirectly) the expression of all terminal differentiation features of a neuron (Hobert 2008; Bertrand and Hobert 2010). The paired-like homeodomain transcription factor UNC-42 has been proposed to function as the terminal selector in the developmental specification of the ASH polymodal nociceptors in C. elegans (Baran et al. 1999). Consistent with the role of a terminal selector, unc-42(e419) loss-of-function mutants were shown to lack the expression of the putative chemoreceptor genes sra-6 and srb-6 specifically in the ASHs, while, for example, srb-6 expression in the ADL and ADF sensory neurons was unaffected (Baran et al. 1999). unc-42(e419) mutants also lack ASH expression of several other terminal identity markers: three Gα encoding genes (gpa-11, gpa-13 and gpa-15), the neuropeptide encoded by flp-21, and the eat-4 vesicular glutamate transporter (Serrano-Saiz et al. 2013). Furthermore, loss of UNC-42 function disrupted behavioral responses to high osmolarity and nose touch (Baran et al. 1999), which are both detected primarily by the ASHs (Bargmann et al. 1990; Kaplan and Horvitz 1993). However, the severity of the nose touch defect was likely due to developmental defects in both the ASH sensory neurons as well as some downstream interneurons, where altered glutamate receptor expression accompanies loss of UNC-42 (Baran et al. 1999; Brockie et al. 2001).
| Identity marker | Expression affected in unc-42(gk598) mutant background | ||
| ASH nociceptive neuron | High osmolarity detection | osm-10 | YES |
| GPCR | sra-6 sri-14 srd-33 srd-44 srd-10 srz-1 |
YES* YES YES YES YES (partial) NO |
|
| Transcription factor | hlh-2 ceh-23 nhr-79 |
YES YES NO |
|
| Neuropeptide | nlp-15 nlp-3 |
YES NO |
|
| TRP channel | osm-9 ocr-2 |
NO NO |
|
| Axon guidance | unc-40 sax-3 |
NO NO |
|
Table 1: Summary of the effect of unc-42(gk598) on ASH terminal marker gene expression
*sra-6 expression in ASH was previously reported to be affected by unc-42(e419) (Baran et al. 1999).
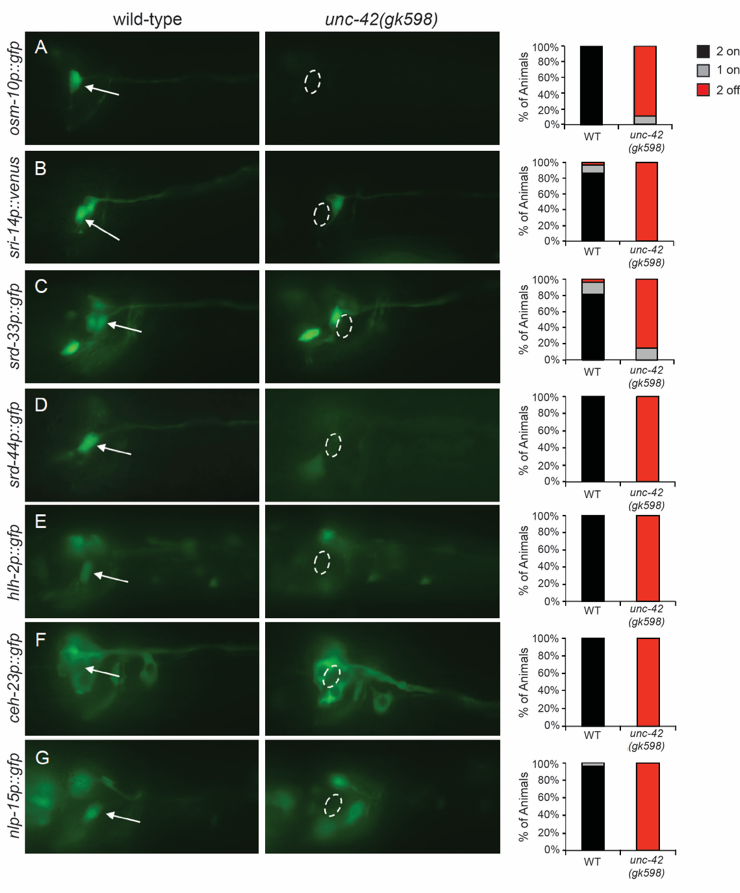
Using the unc-42(gk598) allele, we confirmed the role of UNC-42 in regulating ASH terminal markers, and identified seven additional genes whose ASH expression strongly depends upon UNC-42 (Figure 1 and Table 1). These genes encode OSM-10, additional predicted GPCRs, transcription factors and a neuropeptide. However, UNC-42 is unlikely to function as the sole terminal selector in ASH. For example, srd-10 expression is only partially affected in the unc-42(gk598) mutants, with only ~20% of animals losing expression in one ASH (image not shown). In addition, ASH expression of some genes (srz-1, nhr-79, nlp-3, osm-9, ocr-2, unc-40 and sax-3) was found to be unaffected in unc-42(gk598) animals (Table 1). We note that several of these are either expressed broadly throughout the nervous system, or function in several sensory neurons. As such, they may not necessarily be constituents of the terminally differentiated gene battery of the ASHs. Combined, UNC-42 broadly regulates the expression of ASH identity markers, although additional transcription factors are likely to also contribute.
Acknowledgments
We thank Anne Hart, Takaaki Hirotsu and Piali Sengupta for reagents and Oliver Hobert for valuable discussions.
Reagents
DiD was purchased from Molecular Probes (Invitrogen).
The VC1444 unc-42(gk598) strain was generated by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which is part of the International C. elegans Gene Knockout Consortium. The gk598 allele contains a 1430 basepair deletion (898 basepairs of 5’ UTR sequence, exon 1 and 481 basepairs of intron 1). VC1444 was outcrossed 6x to N2 to generate FG498 for use in this study. Some of the strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded in part by the National Institutes of Health – National Center for Research Resources.
Strains used in this study include: N2 Bristol wild-type, FG498 unc-42(gk598), CX3465 kyIs39 [sra-6::gfp + lin-15(+)], HA1695 rtIs27 [osm-10p::gfp], PY5163 [oyEx srd-44p::gfp, coelRFP] line 1, NK885 unc-119(ed4);qyIs174 [hlh-2p::gfp::hlh-2 + unc-119(+)], CX2565 kyIs4 lin-15A(n765) [ceh-23p::unc-76::gfp + lin-15(+)], RW10748 unc-119(ed3); zuIs178; stIs10024; stIs10549 [stIs10549 = nhr-79::H1-wCherry + unc-119(+)], HA357 rtEx251 [nlp-15p::gfp + lin-15(+)]; lin-15B lin-15A(n765), AL132 icIs132 [unc-40::gfp], HA341 lin-15; rtEx235 [nlp-3::gfp], LX990 lin-15B lin-15A(n765); vsEx494 [ocr-2p::gfp::ocr-2 3’utr + lin-15(+)], CX3716lin-15B lin-15A(n765) kyIs141 [osm-9::gfp5 + lin-15(+)], IC692 quEx162 [sax-3p::gfp + (pRF4) rol-6]. Transgenes were examined in the wild-type andunc-42(gk598) background. For the generation of extrachromosomal arrays, germline transformations were performed as previously described (Mello et al. 1991). Plasmids injected for analysis include: sri-14p::venus, pFG258 srd-33p::gfp and pFG233 srd-10p::gfp. Transgenic lines were examined in the N2 wild-type background, and at least one array was chosen to be crossed into the unc-42(gk598) background for comparison. Strains generated in our lab for this study have not been sent to the CGC.
References
Funding
This work was supported by the National Science Foundation (grant 1351649 to DMF).
Reviewed By
Jagan SrinivasanHistory
Received: April 29, 2019Accepted: May 10, 2019
Published: May 15, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Wood, JF; Ferkey, DM (2019). unc-42 regulates the expression of ASH terminal fate markers. microPublication Biology. 10.17912/micropub.biology.000114.Download: RIS BibTeX