Description
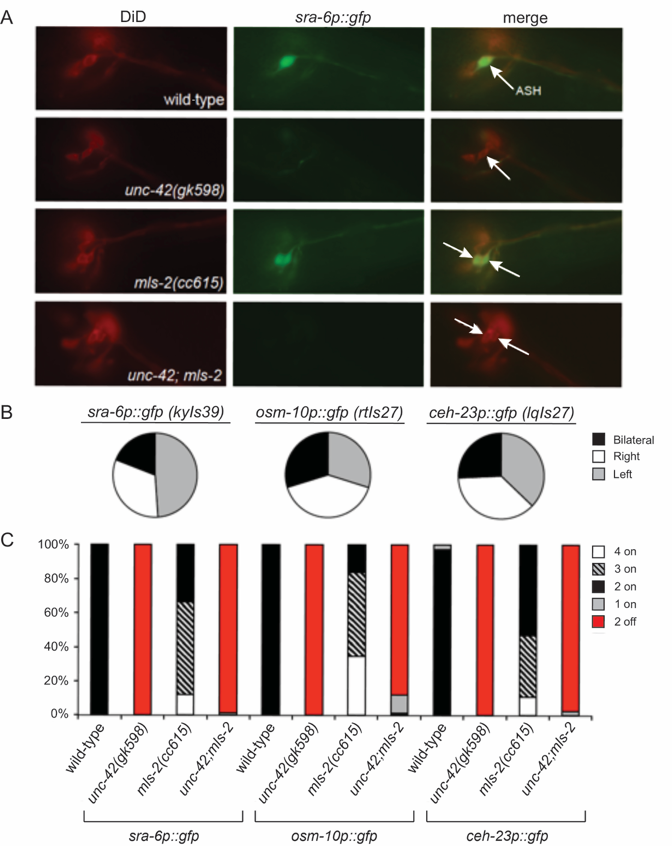
The HMX/NKS homeodomain transcription factor MLS-2 is required to initiate the expression of the AWC terminal selector ceh-36, and as such the AWCs of mls-2 loss-of-function mutant animals fail to express downstream AWC-specific terminal differentiation genes (Kim et al., 2010). Interestingly, loss of mls-2 function was also shown to result in ectopic expression of ASH markers (sra-6p::gfp and osm-10p::gfp) in at least one neuron in a large percentage of mls-2 mutant animals (Kim et al., 2010). The cells that ectopically express the ASH markers are adjacent to the native ASHs and, like native ASH neurons, dye-fill with lipophilic dyes such as DiD (Perkins et al., 1986; Kim et al., 2010).
Since ASH expression of both sra-6p::gfp (Baran et al., 1999; Wood and Ferkey, 2019) and osm-10p::gfp (Wood and Ferkey, 2019), as well as ceh-23p::gfp (Wood and Ferkey, 2019), depends upon the paired-like homeodomain transcription factor UNC-42, we assessed whether the ectopic expression of these ASH markers requires UNC-42 function as well. We first examined the expression pattern of stably integrated reporters for sra-6p::gfp, osm-10p::gfp and ceh-23p::gfp in mls-2(cc615) loss-of-function mutant animals. In addition to being expressed in the native ASH neurons, for each transgene we confirmed ectopic marker expression unilaterally or bilaterally in the ectopic ASH-like cells of mls-2(cc615) mutant animals, which were identified by dye-filling (Figure 1). We note that there was no obvious directional bias as to which side of the bilateral ASH-like pair the unilateral ectopic expression arose (Figure 1B). We found that both native and ectopic expression of all three ASH markers was lost in the unc-42(gk598);mls-2(cc615) double mutants, although the native and ectopic cells retained dye-filling capacity (Figure 1A, C). Thus, the ectopic expression of these ASH markers in the absence of MLS-2 function depends upon UNC-42, as native ASH expression does (Baran et al., 1999; Wood and Ferkey, 2019).
Reagents
DiD was purchased from Molecular Probes (Invitrogen).
The VC1444 unc-42(gk598) strains was generated by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which is part of the International C. elegans Gene Knockout Consortium. The gk598 allele contains a 1430 basepair deletion (898 basepairs of 5’ UTR sequence, exon 1 and 481 basepairs of intron 1). VC1444 was outcrossed 6x to N2 to generate FG498 (Wood and Ferkey, 2019).
Strains used in this study include: N2 Bristol wild-type, FG498 unc-42(gk598), LW227 mls-2(cc615), FG746 unc-42(gk598);mls-2(cc615), CX3465 kyIs39 [sra-6::gfp + lin-15(+)], FG750 unc-42(gk598);kyIs39, FG749 mls-2(cc615);kyIs39, FG748 unc-42(gk598);mls-2(cc615);kyIs39, HA1695 rtIs27 [osm-10p::gfp], FG573 unc-42(gk598);rtIs27,FG745 mls-2(cc615);rtIs27,FG747unc-42(gk598);mls-2(cc615);rtIs27, LE732 lqIs27 [ceh-23::gfp + lin-15(+)], FG839 unc-42(gk598);lqIs27, FG840 mls-2(cc615);lqIs27,FG841 unc-42(gk598);mls-2(cc615);lqIs27. Some of the strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded in part by the National Institutes of Health – National Center for Research Resources. Strains generated in our lab for this study have not been sent to the CGC, but are available by request.
Acknowledgments
We thank Paul Cullen, Todd Hennessey, Jerry Koudelka and Oliver Hobert for valuable discussions.
References
Funding
This work was supported by the National Science Foundation (grant 1351649 to DMF).
Reviewed By
Renee BaranHistory
Received: May 16, 2019Accepted: June 7, 2019
Published: June 12, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Wood, JF; Ferkey, DM (2019). UNC-42 function is required for the ectopic expression of ASH markers in mls-2 mutant animals. microPublication Biology. 10.17912/micropub.biology.000116.Download: RIS BibTeX