Department of Biological and Allied Health Sciences, Ohio Northern University
Department of Biology, Albion College
Biology Department, University of Detroit Mercy
Description
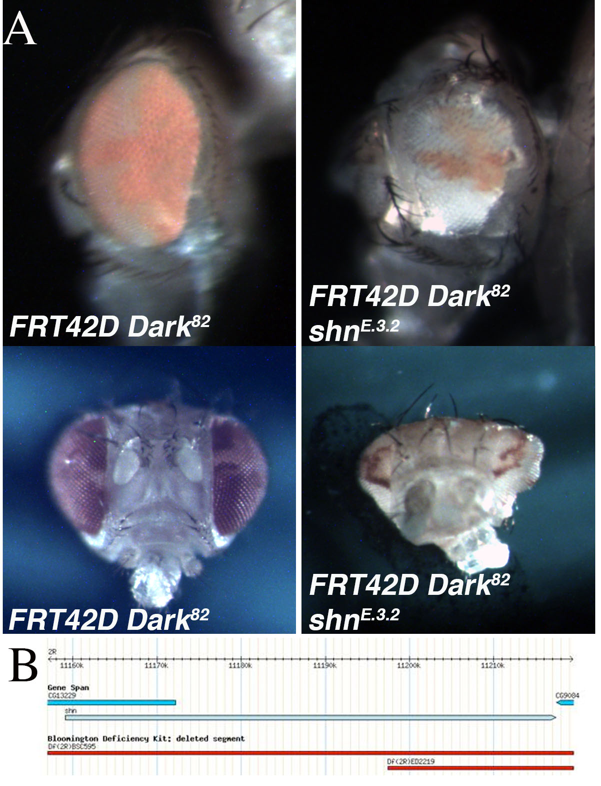
An EMS screen was conducted utilizing the Flp/FRT system to identify mutations that lead to phenotypic alterations in the size of the eye, the ratio of mutant to wild type tissue (red over white), or the developmental patterning of the mosaic eye. This screen was completed in the genetic background of blocked apoptosis in the homozygous mutant cells to identify conditional regulators of cell growth and eye development (Kagey et al., 2012). The block in apoptosis in the mosaic mutant tissue was achieved by using the FRT42D Dark82chromosome, which retains the w+mC(pigmentation), as a starting point for the EMS mutagenesis (Akdemir et al., 2006). One of the mutants identified was mutant E.3.2. The mutant mosaic phenotype, generated from the cross FRT42D Dark82 E.3.2 XEy>Flp; FRT42D, resulted in a range of phenotypes. This included, gross eye pattern disruption, abnormal shape, and antennal overgrowth (see bottom image) when compared to the FRT42D Dark82 mosaic controls (Figure 1A). The control mosaic phenotype had a characteristic 60:40 red:white ratio as compared to the mutant mosaic phenotype of approximately 30:70 red:white ratio. This indicates a reduction in mutant tissue, but included abnormal cranial, eye, and antennal development (Figure 1A).
The genetic mapping of the location of E.3.2 on 2R was completed by three independent groups of undergraduate researchers at Nevada State College, Albion College, and Ohio Northern University as part of the Fly-CURE consortium (Bieser et al., 2018, Stamm et al., 2019). Virgin females from the FRT42D E.3.2 Dark82/CyO stock were mated in series to male flies from the 87 deficiency stocks that comprise the Bloomington Stock Center 2R Deficiency Kit (only stocks distal to the FRT42D site were used for mapping) (Cook et al., 2012). Mutant E.3.2 failed to complement deficiencies Df(2R)BSC595 (2R:10,385,967..11,288,578), Df(2R)ED2155 (2R:10,894,096..11,397,442), and Df(2R)ED2219 (2R:11,197,412..11,665,391), while complementing the flanking deficiency Df(2R)Exel6059 (2R:10,874,385..11,186,047). This resulted in the genomic region of 2R:11,186,047..11,665,391 failing to complement (Figure 1B). A lethal allele for the candidate gene Schnurri, shn[1],was crossed independently to E.3.2 to test for complementation. E.3.2 failed to complement a previously defined strong hypomorphic allele, shn[1] (Horsfield et al., 1998), indicating that E.3.2 is a novel shn allele, shnE.3.2. Shn is a zinc-finger transcription factor known to act in complex with Mothers Against Dpp (mad) in response to the BMP-related Decapentaplegic (Dpp) signaling pathway (Dai et al., 2000; Udagawa et al., 2000). Dpp, a member of the TGFβ superfamily, is a Drosophila morphogen critical for directing early embryonic patterning and regulating cell growth. Dpp mutants have shown that Dpp signaling is necessary for proper patterning in the eye-antenna disc in the L2 and L3 wandering larval stages of Drosophila development (Won et al., 2015). The current work is in agreement with previous findings and indicate that perturbations of shn, a downstream target of Dpp, is involved in regulation of cell growth and developmental patterning in vivo.
Reagents
FRT42D Dark82/CyO (Akdemir et al., 2006)
FRT42D Dark82 shnE.3.2/CyO (this manuscript)
Ey>Flp; FRT42D (BDSC 5616)
Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al., 2012)
w1118;Df(2R)BSC595/CyO (BDSC 25428)
w1118;Df(2R)ED2219,P{w[+mW.Scer\FRT.hs3]=3’.RS5+3.3’}ED2219/Sm6a (BDSC 8910)
w1118;Df(2R)ED2155,P{w[+mW.Scer\FRT.hs3]=3’.RS5+3.3’}ED2155/SM6a (BDSC 9344)
w1118;Df(2R)Exel6059,P{w[+mC]=XP-U}Exel6059/CyO (BDSC 7541)
cn[1] shn[1] bw[1] sp[1]/CyO (BDSC 3008)
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
References
Funding
None
Reviewed By
Anonymous and Michael O'ConnorHistory
Received: May 24, 2019Accepted: June 4, 2019
Published: June 5, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Bieser, K; Sanford, JS; Saville, K; Arreola, KF; Ayres, ZT; Basulto, D; Benito, S; Breen, CJ; Brix, JA; Brown, N; Burton, KK; Chadwick, TM; Chen, M; Chu, K; Corbett, BL; Dill, Z; Faughender, MA; Hickey, AD; Julia, JS; Kelty, SS; Jobs, BB; Krason, BA; Lam, B; McCullough, CL; McEwen, BR; McKenzie, JL; McQuinn, KR; Moritz, CM; Myers, KE; Naugle, EM; Nutter, AM; O'Conke, DQ; O'Grondik, MT; Patel, KB; Rudowski, SM; Sberna, EN; Stall, GM; Steiner, TL; Tanriverdi, E; Torres Patarroyo, N; Traster, VL; Tsai, LP; Valenti, AJ; Villegas, MM; Voors, SM; Watson, KK; Wright, ME; Kagey, JD (2019). Genetic mapping of shnE.3.2 in Drosophila melanogaster. microPublication Biology. 10.17912/micropub.biology.000118.Download: RIS BibTeX