Department of Cell Biology, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan
Description
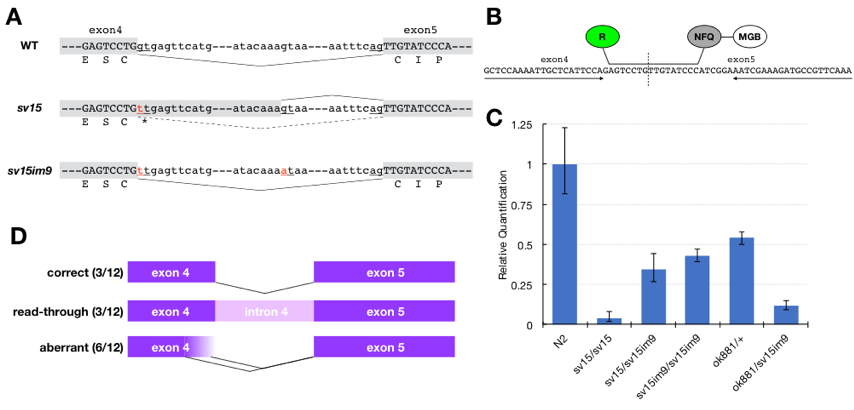
TSP-15 is one of the 21 tetraspanins in Caenorhabditis elegans and is essential for exoskeletal (cuticle) development. A reduction in the function of TSP-15 results in a dumpy (Dpy) and/or blistered (Bli) phenotype (Moribe et al. 2004), and TSP-15 null mutants exhibit embryonic lethality (Moribe et al. 2012). sv15 is a hypomorphic allele of tsp-15, featuring a splicing error mutation in tsp-15’s fourth intron. The mutation results in the immediate production of a stop codon (Fig. 1A), and the resulting truncated form of TSP-15 protein has no function. sv15 mutants are viable despite severe cuticle deficiencies since some correctly spliced transcripts are produced (Moribe et al. 2004).
To further understand the functions of TSP-15 in vivo, we screened genetic suppressor mutants of tsp-15(sv15). tsp-15(sv15) worms were mutagenized with 50 mM ethyl methanesulfonate (EMS) for 4 h and F2 progeny were screened for wild-type appearance. Nine suppressor alleles (im1 to im9) were isolated from a nonclonal screen from 15,000 haploid genomes. One dominant and eight recessive suppressor alleles were included among them. The alleles were mapped by single-nucleotide polymorphism (SNP) mapping using Hawaiian variant CB4856 and characterized by DNA sequence analysis. Mutations were confirmed by complementation test, rescue by DNA transformation, and RNAi analysis. We identified one dominant allele as described here and four of eight recessive alleles, which will be described elsewhere.
The dominant allele im9 was revealed to feature an intragenic mutation in intron 4 of tsp-15 (Fig. 1A). sv15/sv15im9 showed the wild-type appearance with a very mild Dpy phenotype, and the appearance of sv15im9/sv15im9 homozygotes almost matched of the wild-type. im9 mutation abolished a cryptic splice donor site, which is used in sv15, suggesting that the original splice site was possibly reused in sv15im9 mutants. To test this hypothesis, we performed quantitative RT-PCR (qRT-PCR). To detect the correct tsp-15 transcript, we designed a TaqMan MGB probe specific for the border between exons 4 and 5 (Fig. 1B). The tsp-15 primer set and TaqMan probe were designed by Primer Express software (Applied Biosystems). Total RNA and first-strand cDNA were prepared as described previously (Moribe et al. 2004). TaqMan real-time PCR was performed and analyzed using the ABI 7500 system (Applied Biosystems). RNA polymerase II subunit ama-1 was used as an internal control to normalize the tsp-15 relative value, and its primers and probe were purchased from TaqMan Gene Expression Assays (Assay ID Ce02462726_m1). The PCR reaction was performed using TaqMan Universal PCR Master Mix (Applied Biosystems), in accordance with the manufacturer’s protocol. Because the amplification efficiencies of tsp-15 and ama-1 were almost equal, we calculated the relative amount of tsp-15 by the comparative CT method. The relative value was determined from the mean of duplicate experiments. The results showed that the correct transcript was produced from the sv15im9 allele at a high level. sv15im9 homozygotes were comparable to tsp-15 null heterozygotes (ok881/+), which showed the wild-type appearance, in terms of the expression level of the correct transcript of tsp-15 (Fig. 1C). This suggested that the tsp-15 expression level in sv15im9 was sufficient to restore exoskeletal development. In addition, we also sequenced tsp-15 cDNA subclones isolated from sv15im9 by RT-PCR, and confirmed the presence of correctly spliced transcripts (Fig. 1D).
The C. elegans splice site consensus sequences match those of vertebrates; C. elegans introns also obey the GU-AG rule (Blumenthal and Steward 1997). The splicing that occurred in sv15im9 mutants did not conform to this rule, but several instances in which the splice site did not always follow the rule have already been reported. Intron recognition involves the A+U richness of introns, which is probably significant for recognition of 5′ splice sites by U1 snRNP (Blumenthal and Steward 1997).
Reagents
Strains:
OB83 tsp-15(sv15/sv15im9)
OB119 tsp-15(sv15im9/sv15im9)
OB176 tsp-15(ok881/sv15im9)
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Edanz (www.edanzediting.co.jp) for editing the English text of a draft of this manuscript.
References
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP26440107 to HM and JP26250033 to EM.
Reviewed By
AnonymousHistory
Received: June 11, 2019Accepted: July 18, 2019
Published: July 23, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Moribe, H; Mekada, E (2019). Isolation of intragenic suppressor of tsp-15-splicing mutant in Caenorhabditis elegans. microPublication Biology. 10.17912/micropub.biology.000126.Download: RIS BibTeX