Present address: McArdle Laboratory for Cancer Research, University of Wisconsin, Madison, WI
Description
Mutations in the C. elegans gene srf-6 result in an alteration in surface composition detected with monoclonal antibody (mAb), which is specific for the wild-type L1 surface. In srf-6 mutants, all larval stages display the mAb epitope (Hemmer et al., 1991; Grenache et al., 1996). This suggests that srf-6 might be a component of a genetic switch that operates in wild-type to turn off expression of the epitope after the L1 stage. We were therefore interested in identifying the srf-6 DNA sequence.
Genomic DNA was isolated from srf-6(yj13), srf-6(yj41), and srf-6(yj15) worms grown on plates, using a Gentra Puregene Kit. Whole genome sequencing of DNA was carried out at the Caltech Genomics Facility. DNA sequences from each mutant were analyzed using the CloudMap workflow (Minevich et al., 2012) on the Galaxy Internet platform (Afgan et al., 2018). After aligning sequences with the C. elegans reference genome, variants were called using the GATK Unified Genotyper. To remove variants that were shared in common among all three srf-6 mutants, the GATK Select Variants plugin was used to subtract the union of two mutant variant sets from the third one. This was repeated for all three pairwise subtractions to produce sets of mutant-specific variants. The effect of each variant on protein structure was predicted, and the resulting sets of predicted changes were merged and subjected to the Cloudmap in silico complementation tool, which sorts variants according to the genetic location and C. elegans gene in which they occur.
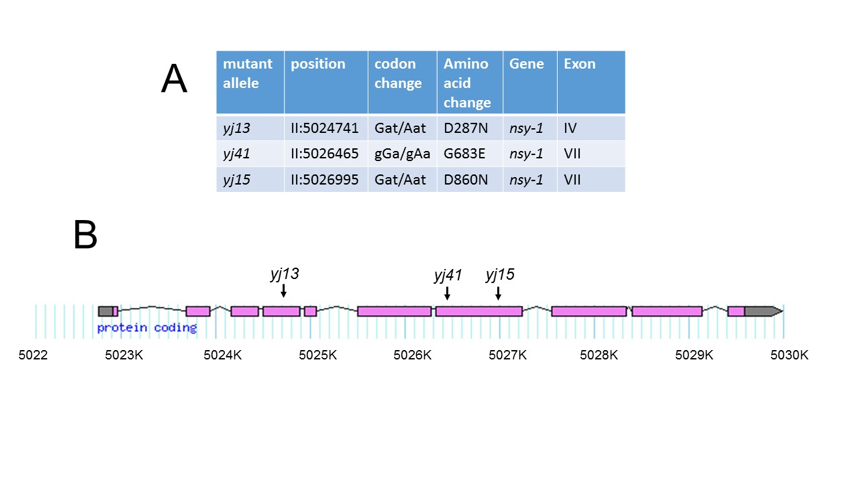
Results of the in silico complementation analysis are shown in Figure 1A. Each strain contained a G to A transition in a chromosome II gene sequence, F59A6.1, which corresponds to the gene nsy-1. This result is consistent with the mutations having been induced by ethylmethanesulfonate (Hemmer et al., 1991), and is also consistent with the previous assignment of srf-6(yj13) to linkage group II (Grenache et al., 1996). Each mutation resulted in a non-conservative amino acid substitution in the encoded protein, including an aspartic acid changed to asparagine in srf-6(yj13)and srf-6(yj15), and a glycine changed to glutamic acid in srf-6(yj41). Positions of these mutations are shown in Figure 1B. Mutations carried by srf-6(yj15)and srf-6(yj41) were located in exon 7, which contains the kinase catalytic domain (Sagasti et al., 2001). The mutation carried by srf-6(yj13) was located in exon 4. All three mutations altered amino acids that are conserved between NSY-1 and its closest human homolog, ASK-1 (Ichijo et al., 1997).
Reagents
C. elegans strains
AT18 srf-6(yj13) II
AT24 srf-6(yj15) II
AT25 srf-6(yj41) II
Strains will be submitted to the Caenorhabditis Genetics Center.
Acknowledgments
This work was supported by DNA sequencing carried out by Igor Antoshechin at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The authors thank Mark Alkema for advice on genomic DNA isolation, colleagues Elizabeth Ryder and Jagan Srinivasan for comments on the manuscript, and Chris Link for suggesting that we try whole genome resequencing to identify srf-6.
References
Funding
This work was partially funded by the Office of the Dean of Arts and Sciences, Worcester Polytechnic Institute, Worcester, MA.
Reviewed By
Maria Gravato-NobreHistory
Received: June 18, 2019Accepted: June 27, 2019
Published: July 4, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Van Sciver, ND; Pulkowski, JO; Politz, SM (2019). Three C. elegans srf-6 mutants carry nsy-1 mutations (srf-6 is nsy-1 I). microPublication Biology. 10.17912/micropub.biology.000127.Download: RIS BibTeX