Department of Mathematical Sciences, Worcester Polytechnic Institute, Worcester, MA
Department of Biology and Biotechnology, Worcester Polytechnic Institute, Worcester, MA
Description
In previous work, we showed that three different srf-6 mutants carry mutations in gene nsy-1 (Van Sciver et al., 2019), and that a srf-6 mutant shows the 2AWCON phenotype characteristic of nsy-1 mutants. We also showed that srf-6(yj13) fails to complement nsy-1(ok593) (Honzel et al., 2019). We concluded that srf-6 and nsy-1 are the same gene.
C. elegans gene nsy-1 encodes a MAP kinase kinase kinase (MAPKKK) that carries out the first step in a C. elegans MAP kinase pathway (Kim et al., 2002, Sagasti et al., 2001, Tanaka-Hino 2002). The pathway culminates in activation of a MAP kinase that is homologous to the human p38 MAP kinase (Kim et al., 2002). This pathway is involved in at least three biological processes in C. elegans, i.e., immune responses to infection by pathogenic microbes (Kim et al., 2002), upregulation of serotonin biosynthesis in the ADF chemosensory neurons in response to pathogen exposure (Shivers et al., 2009, Zhang, Lu, and Bargmann 2005), and determination of asymmetric cell fate in the AWC chemosensory neuron pair (Sagasti et al., 2001).
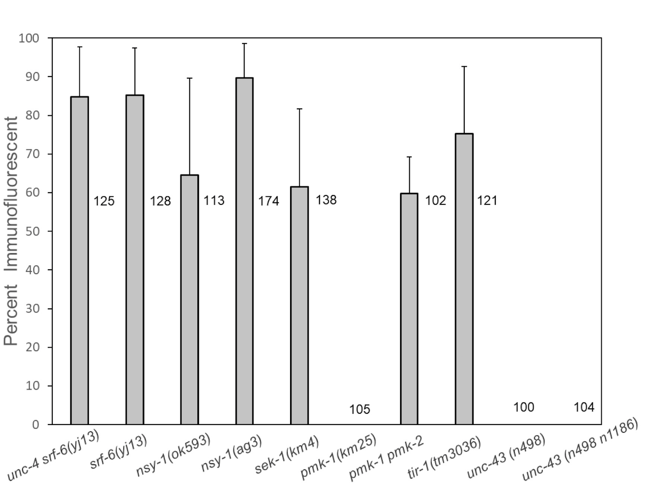
C. elegans srf-6 mutants were originally identified by antibody staining at stages L2-L4 with mAb M37, which stains only the L1 stage in wild type C. elegans under normal growth conditions (Hemmer et al., 1991, Grenache et al., 1996). To determine whether loss of function in the C. elegans p38 pathway also results in staining at stages L2-L4, we tested nsy-1 (MAPKKK), sek-1 (MAPKK) and pmk-1 (MAPK) mutants for mAb M37 staining at these later larval stages. Results are shown in Figure 1.
In these experiments, nsy-1(ok593) and nsy-1(ag3) also showed staining of L2-L4 stages (Figure 1). Staining of sek-1(km4) was also positive. The Toll-interleukin-receptor-like protein TIR-1 is required upstream of nsy-1 for both immune responses and determination of AWC fate (Couillault et al., 2004, Chuang and Bargmann 2005). Positive antibody staining was also observed in tir-1(tm3036) mutants.
It was surprising that no L2-L4 larvae of a mutant strain carrying pmk-1(km25) stained (out of a total of 105 worms in 4 biological replicates), because PMK-1 is required as the MAP kinase for immune responses (Kim et al., 2002). However, a requirement for PMK-1 has not been demonstrated for determination of AWC cell fate (Pagano, Kingston, and Kim 2015). It has been shown that function of the TIR-1 NSY-1 SEK-1 pathway is required for determination of AWC fate (Chuang and Bargmann 2005), and that PMK-1 and a second MAP kinase, PMK-2, function redundantly for determination of AWC cell fate (Pagano, Kingston, and Kim 2015). We therefore tested a pmk-2 pmk-1 double mutant for M37 antibody staining. In contrast to pmk-1, the pmk-1 pmk-2 double mutant showed staining at a level similar to that of sek-1(km4).
The unc-43-encoded calcium dependent kinase CaMKII is required for upstream activation of the TIR-1 NSY-1 SEK-1 cascade in the AWC neuron, and the level of unc-43 activity in AWC can determine whether the cell develops the AWCONor the AWCOFF fate (Troemel, Sagasti, and Bargmann 1999). We tested the mutant unc-43(n498gf), which shows a 2AWCOFFphenotype, and the 2AWCONmutant unc-43(n498n1186lf) for M37 antibody staining (Troemel, Sagasti and Bargmann 1999). Neither of these mutants stained with mAb M37.
Reagents
C. elegans Strains
AT18 srf-6(yj13) II
VC390 nsy-1(ok593) II
AU3 nsy-1(ag3) II
KU4 sek-1(km4) IV
KU25 pmk-1(km25) II
ZD1006 pmk-2(qd279qd171) pmk-1(km25) II
IG685 tir-1 (tm3036) III
MT1092 unc-43(n498) IV
MT2605 unc-43(n498) unc-43(n1186) IV
All strains are available from the Caenorhabditis Genetics Center, except for AT18, which will be sent to the CGC.
Acknowledgments
The authors thank members of the Politz lab for discussion.
References
Funding
Partial funding for this work was provided by a grant from the office of the Dean of Arts and Sciences, Worcester Polytechnic Institute.
Reviewed By
Maria Gravato-NobreHistory
Received: June 18, 2019Accepted: June 27, 2019
Published: July 4, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Foley, SJ; Wu, Z; Politz, SM (2019). A C. elegans MAP kinase pathway is required for wild-type display of an L1-specific surface antigen (srf-6 is nsy-1 III). microPublication Biology. 10.17912/micropub.biology.000129.Download: RIS BibTeX