Current address: Department of Molecular, Cellular, and Developmental Biology, University of Colorado Boulder, Boulder, CO 80303
Description
KQT-3 is the C. elegans ortholog of human KCNQ1, a voltage-gated potassium channel that is implicated in multiple types of Long Q-T cardiac arrhythmias including Jervell and Lange-Nielsen syndrome and Romano-Ward syndrome. In order to assess the possibility of modeling these syndromes in C. elegans, we obtained the tm542 deletion allele, which had previously been associated with changes to the defecation cycle (Nehrke et al., 2008, Kwan et al. 2008). Any visible, easily-scored phenotype would be an indication that we could move forward with creating patient-specific missense alleles by CRISPR.
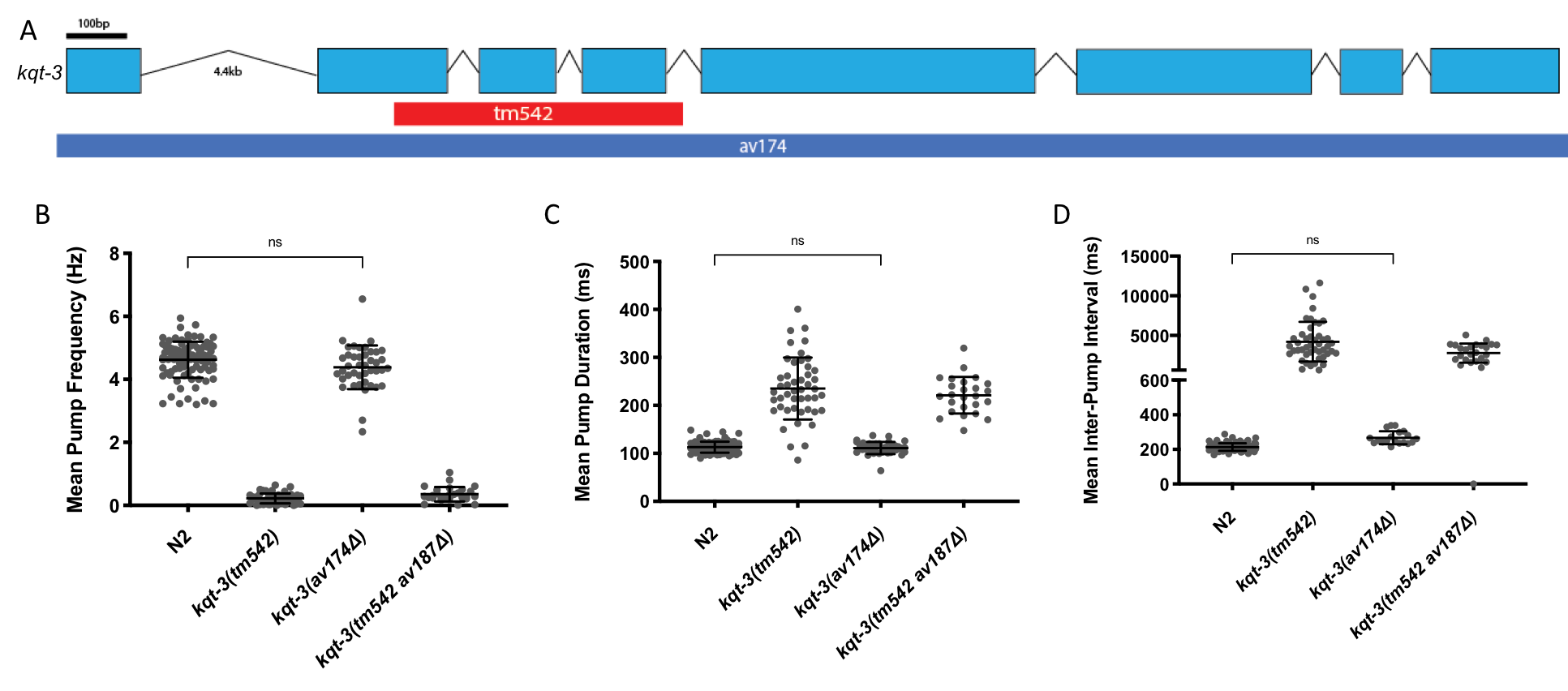
The tm542 deletion, a 1001bp deletion generated by chemical mutagenesis that disrupts exons 2-4, appeared to cause a dramatic reduction in pharyngeal pump frequency (Fig 1B). This was an indication that mutations to kqt-3 might be a reasonable way to model Long Q-T arrhythmias in worms. When three patient missense mutations and two smaller deletions yielded only very subtle changes to pharyngeal pumping, we became increasingly suspicious of the tm542 deletion strain. This prompted the creation of a full-length, 9004bp deletion allele by CRISPR that starts 69bp upstream of the start codon, ends 66bp after the stop codon, and inserts stop codons in every frame. Surprisingly, complete deletion of this 621 amino acid protein led to virtually no change in pharyngeal pumping (Fig 1B-D).
In order to confirm that the pharyngeal pumping phenotype in the kqt-3(tm542) strain was due to extraneous mutations and not a dominant negative effect, we created the full-length deletion of kqt-3 by CRISPR in the tm542 strain. This kqt-3(tm542av187Δ) deletion strain phenocopied the original tm542 deletion (Fig 1B-D), indicating that other mutations besides the deletion in kqt-3 are responsible for the lack of pharyngeal pumping in the tm542 strain.
This study should serve as a cautionary tale for any lab studying a deletion allele that was generated by chemical mutagenesis; the strain likely contains numerous other mutations. We now routinely generate a full gene deletion by CRISPR as part of any genetic study.
Reagents
Strains
FX542 kqt-3(tm542) II
AG451 kqt-3(av174∆) II
AG479 kqt-3(tm542av187∆) II
Genotyping
Oligos used for genotyping tm542: expect sizes 1342bp WT and 340bp tm542.
F: 5’-ttccggtaagaaacttctgg
R: 5’-cccaaaactccaccaattagc
Oligos used for genotyping av174∆ and av187∆: one internal pair and one external pair; expect sizes 716bp WT from internal reaction and 486bp full gene deletion from external reaction.
F internal: 5’-cgccacaaatttttgtgtcc
R internal: 5’-ggtcagaaatgttgcggaat
F external: 5’-gctcattctctctcacaccac
R external: 5’-cccaaatcatgttgacagcgag
Electropharyngeograms
15 gravid hermaphrodites were placed on OP50-seeded MYOB plates for 6 hours to establish a synchronized population. After 3 days the population was washed off with M9, washed 3x, then incubated in 10mM 5-HT for at least 20 minutes. Pharyngeal pumping was then recorded using the NemaMetrix ScreenChip system (Eugene, OR). All strains were recorded on at least two separate days. Outliers were identified by ROUT with Q=0.5%. Strains were compared by one-way ANOVA with Tukey’s multiple comparison test.
CRISPR/Cas9
The av174 and av187 deletions were created using clone-free CRISPR-mediated gene editing (Paix et al. 2015) with dpy-10 co-CRISPR (Arribere et al. 2014). The deletion was generated using crRNAs at both the 5’ and 3’ ends of kqt-3 (5’: 5’-actagtagtaatatattaaa; 3’: 5’-gaccacgacgcatcaacagc) and a single-stranded repair template containing an insertion of stop codons in every frame (TAGATAAATGA) and 35bp of homology on both sides. This mix was injected into N2 to obtain kqt-3(av174∆) and into kqt-3(tm542) to obtain kqt-3(tm542av187∆).
Acknowledgments
We thank Srinath Seshadri for assistance in performing electropharyngeograms and the National Bioresource Project (Japan) for providing strains.
References
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute of Diabetes, Digestive and Kidney Diseases.
Reviewed By
Nicole LiachkoHistory
Received: July 11, 2019Accepted: July 15, 2019
Published: July 25, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Bauer, R; Nebenfuehr, B; Golden, A (2019). Resolving a contradiction: full gene deletion of kqt-3 by CRISPR does not lead to pharyngeal pumping defects. microPublication Biology. 10.17912/micropub.biology.000137.Download: RIS BibTeX