Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, 100101, China
Neurobiology Section, Division of Biological Sciences, University of California, San Diego, CA92093
Description
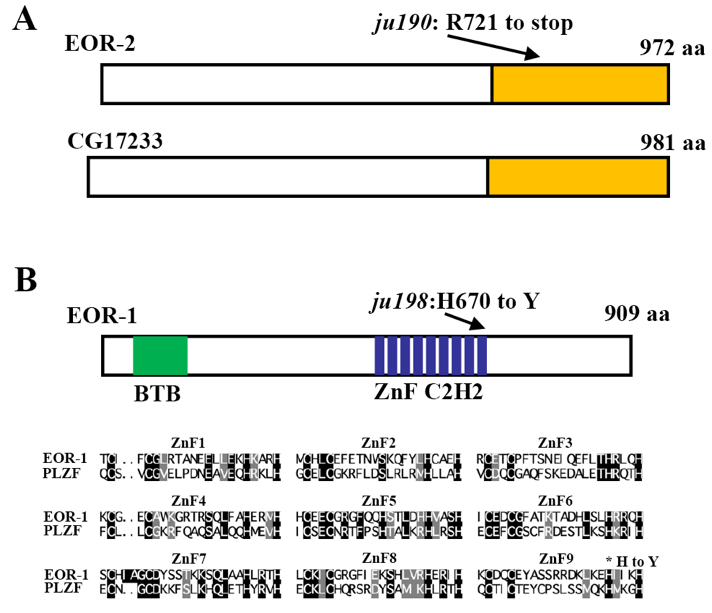
In a genetic screen for genes affecting RMED/V neuron specification, we isolated two mutants, ju190 and ju198 (Huang et al., 2002; Huang et al., 2004; Huang and Jin, 2019). We mapped ju190 to the X chromosome, a region covered by three cosmids (H01A20, C44H4 and F54E4), between unc-9 and unc-3, using the snip-SNP mapping strategy. The novel conserved nuclear transcription factor eor-2 is contained in the cosmid C44H4, and eor-2(cs42) mutant animals exhibit similar behavior defects as ju190 (Rocheleau et al., 2002). We introduced Punc-25GFP into eor-2(cs42), a null allele, and found no expression in RMED/V cell, as for ju190 (Huang and Jin, 2019). DNA sequencing analysis of the eor-2 genomic DNA from homozygous ju190 animals identified a C to T nucleotide transition that results in an Opal stop at Arg721, in the conserved C-terminal domain (Figure 1A). Therefore, the RMED/V defects in ju190 arise from a complete loss of EOR-2 function.
We mapped ju198 to chromosome IV in a region near eor-1. EOR-1 is a functional binding partner of EOR-2 (Howard and Sundaram, 2002; Howell et al., 2010). The phenotypic similarities between eor-1 and eor-2 and between ju198 and ju190 led us to suspect that ju198 might be an allele of eor-1. Indeed, we found that eor-1(cs28), a null allele, failed to complement ju198 for the RMED/V phenotypes (Huang and Jin, 2019). eor-1 encodes a C. elegans ortholog of mammalian promyelocytic leukemia zinc finger protein (PLZF) with a BTB domain and nine C2H2 zinc fingers (Figure 1B) (Rocheleau et al., 2002). We found that ju198 is a missense mutation changing a conserved histidine to tyrosine in the last zinc finger (Figure 1B). Altogether, our data show that the complete loss of either eor-1 or eor-2 function results in identical differentiation defects in RMED/V neurons.
Reagents
Strains are: CZ2014 eor-1(ju198), juIs76; CZ2006 eor-2(ju190); juIs76
Acknowledgments
Some of the strains used here were obtained from the Caenorhabditis Genetics Center, which is supported by the NIH.
References
Funding
NIH R01 NS 035546
Reviewed By
Oliver HobertHistory
Received: July 1, 2019Accepted: July 18, 2019
Published: July 31, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Huang, X; Jin, Y (2019). New mutants defective in RMED/V neuron specification are alleles of EOR-1 and EOR-2. microPublication Biology. 10.17912/micropub.biology.000139.Download: RIS BibTeX