Description
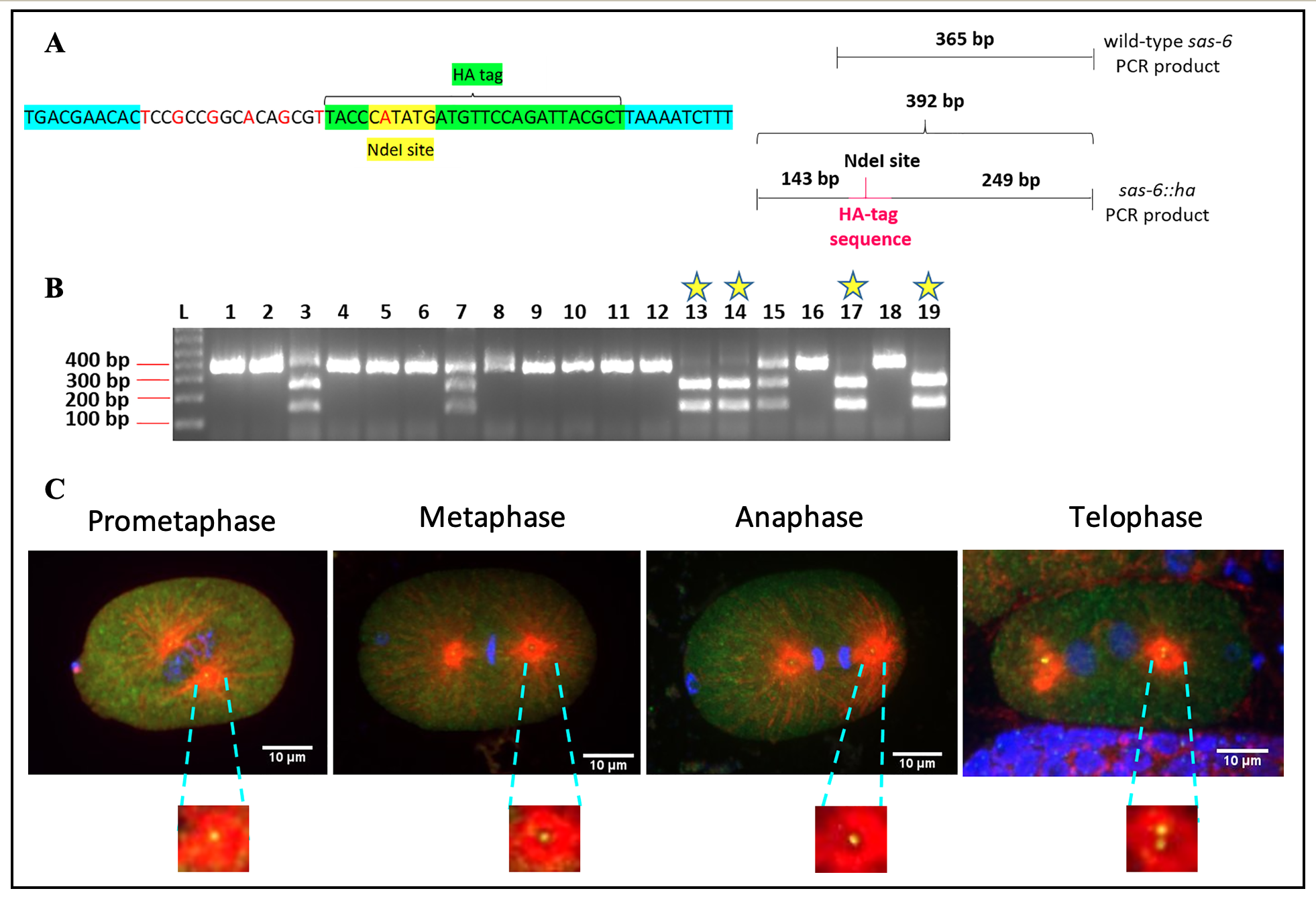
Centrosomes are comprised of a pair of barrel-shaped centrioles that are oriented at right angles to each other, and embedded in electron dense pericentriolar material. The centrosomes mediate spindle assembly and function as basal bodies to promote cilia and flagella formation (reviewed in Pintard and Bowerman, 2019 and Fırat-Karalar and Stearns, 2014). Six core proteins that are required for centriole duplication have been identified in C. elegans. These include the proteins SAS-7, SPD-2, ZYG-1, SAS-6, SAS-5 and SAS-4 (reviewed in Schwarz et al., 2018). The protein SAS-6 is frequently used to mark C. elegans centrosomes as it has been shown to be stably associated with C. elegans centrioles (Dammermann et al., 2004, Liedel et al., 2005, Dammermann et al., 2008 and Balestra et al., 2015). Previous efforts to tag the sas-6 gene with fluorescent tags such as green fluorescent protein (GFP) have, however, resulted in considerable embryonic lethality (Dammermann et al., 2008). One reason for this could be that large tags like GFP interfere with the function of the SAS-6 protein, thereby impairing its activity. Tagging sas-6 with small epitope tags like HA could allow for visualization of endogenous SAS-6 localization without significantly impairing its activity. Although raising antibodies against endogenous C. elegans SAS-6 protein is an attractive alternative to epitope tagging, this is an expensive and time-consuming endeavor. Further, the specificity of antibodies that are raised in this manner cannot be guaranteed. On the other hand, antibodies against short epitope tags such as HA are commercially available, have been well-characterized and are available in a variety of different species (e.g. mouse, rabbit, goat, guinea pig, sheep, etc.) In this study, we have generated a worm strain (IYR001) with the endogenous sas-6 gene ha-tagged using CRISPR/Cas9 editing. Specifically, we have inserted the coding sequence for the HA-tag (9 amino acids YPYDVPDYA) at the C-terminus of the SAS-6 protein. In this CRISPR experiment, we introduced a restriction site for the enzyme NdeI by silent mutation of the HA-tag in our repair template (Figure 1A, left panel). Therefore, all worms that exhibit the incorporation of the NdeI restriction site at the end of the sas-6 gene must have a good chance of being successfully edited to incorporate our supplied repair template. The screening strategy for this CRISPR experiment is depicted in Figure 1A, right panel. Upon performing sas-6 PCRs and digesting these PCR products with NdeI followed by agarose gel electrophoresis, we would expect to detect a single band of about 365 base pairs for wild-type, unedited worms, three bands of approximately 365 base pairs, 249 base pairs and 143 base pairs for heterozygous edited worms and two bands of about 249 base pairs and 143 base pairs respectively for homozygous edited worms (Figure 1A, right panel). As shown in Figure 1B, out of the 19 progeny of positive heterozygotes whose genotypes were analyzed, worm numbers 13, 14, 17 and 19 showed the presence of the homozygous ha-tagedit. We have also confirmed this CRISPR edit by DNA sequencing. At 20°C, C. elegans that are homozygous for the sas-6::ha edit have a slightly reduced average brood size of 240 (n=10) as compared with wild-type C. elegans that have an average brood size of 310 (n=12). However, importantly, sas-6::ha homozygotes do not exhibit any significant embryonic lethality (100% viable (n=10)) as compared with wild-type worms (99% viable (n=12)). We have performed immunostaining on the IYR001 strain with a monoclonal anti-HA antibody to determine the localization of SAS-6::HA in C. elegans embryos. SAS-6::HA displays a stereotypical centrosomal and cytoplasmic localization in 1-cell C. elegans embryos (Figure 1C). We believe that this strain will be a useful tool for C. elegans researchers studying SAS-6 localization and centrosome biogenesis.
Reagents
Immunostaining: Immunostaining of C. elegans embryos was performed as described previously (O’Connell and Golden, 2014) except that TBSBT (TBST with BSA) was used for the blocking and washing steps instead of PBSBT (PBST with BSA). The anti-HA antibody was used at a 1:1000 dilution [Cell Signaling (Catalog # 3724S)] and the anti-tubulin antibody (DM1a) was used at a 1:50 dilution [Santacruz Biotechnology (Catalog # sc-32293)]. Anti-rabbit Alexa fluor 488 (Catalog # A-11034) and anti-mouse Alexa fluor 568 (Catalog # A-11004) secondary antibodies were purchased from Thermo Fisher Scientific and used at a 1:1000 dilution. The embryos were mounted in vectashield containing DAPI [Vector laboratories (Catalog # H-1200)].
CRISPR/Cas9 editing protocol: CRISPR/Cas9 editing was performed as described previously (Paix et al., 2015). The progeny of successfully injected worms were subjected to PCR [MyTaq Redmix, Bioline] followed by restriction digestion with NdeI [New England Biolabs]. Positive edits were identified by agarose gel electrophoresis of digests.
Sequences used for CRISPR/Cas9 editing:
The tracrRNAsequence, dpy-10 crRNA and dpy-10 repair template sequences have been described in Paix et al., 2015.
sas-6 crRNA sequence: 5′ – ATTTTATCGTTGAGCGGGTG – 3′
sas-6::ha repair template sequence:
5'- TATTTTCAAGTAAAGGACAAGAAAAAATCAATAAAAAAGATTTTAAGCGT
AATCTGGAACATCATATGGGTAACGCTGTGCCGGCGGAGTGTTCGTCACA
CTTGAACCAGTAGTCTCGTCGGCGATTAGTTGA – 3'
SAS-6::HA sequencing forward primer: 5′ – CCCCATTCCGTGACAATACA – 3′
SAS-6::HA sequencing reverse primer: 5′ – CCTTACCTCTTGAACTGCC – 3′
Strain name: IYR001
Allele name: sas-6(luv1[sas-6::ha])
The IYR001 strain will be made available upon request.
Imaging: The immunostained embryos were viewed using an Olympus IX83 Yokagawa CSU-X1 spinning disk confocal microscope and images were captured with a prime 95B CMOS camera.
Acknowledgments
We would like to thank the Caenorhabditis Genetics Center for providing us the parental N2 strain which was used as a background to make the IYR001 strain. Many thanks to Nina Peel for critical review and feedback on this microPublication. Special thanks to the Tulsa Summer Undergraduate Research Challenge (TURC) and the Chemistry Summer Undergraduate Research Program (CSURP) at the University of Tulsa for awarding stipends to the students that are involved in this study.
References
Funding
This work was funded by a University of Tulsa start-up grant to Jyoti Iyer. The N2 C. elegans wild isolate strain was provided by the Caenorhabditis Genetics Center, which is supported by NIH Office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
AnonymousHistory
Received: July 16, 2019Accepted: July 25, 2019
Published: August 1, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Bergwell, M; Smith, A; Lakin, H; Slay, R; Iyer, J (2019). Generation of sas-6::ha by CRISPR/Cas9 editing. microPublication Biology. 10.17912/micropub.biology.000141.Download: RIS BibTeX