Description
Insertion of single-copy transgenes is an important tool in C. elegans genome engineering. Following the discovery that the Drosophila mariner element Mos1 could mobilize in the C. elegans germline (Bessereau et al. 2001), tools for efficient Mos1-mediated transgene insertion have been developed and widely adopted (Robert and Bessereau 2010). In parallel, the establishment of effective antibiotic selection in C. elegans offered increased versatility for genetic crosses and the advantage of injecting into phenotypically wild-type animals (Giordano-Santini et al. 2010, Semple et al. 2010).
Reliable, site-specific, single-copy transgene insertion remains a key technique for applications such as synthetic reporters and rapid testing of genetic rescue constructs, especially when germline expression is required. The highly developed Mos1-mediated single-copy insertion (mosSCI) system is a toolkit of choice for this approach (Frokjaer-Jensen et al. 2008, Frokjaer-Jensen et al. 2012). The ttTi5605(mos1)II insertion site in particular has been used for stable germline expression, and plasmids targeting ttTi5605 are also compatible with the ‘universal’ mosSCI insertion sites (Frokjaer-Jensen et al. 2014).
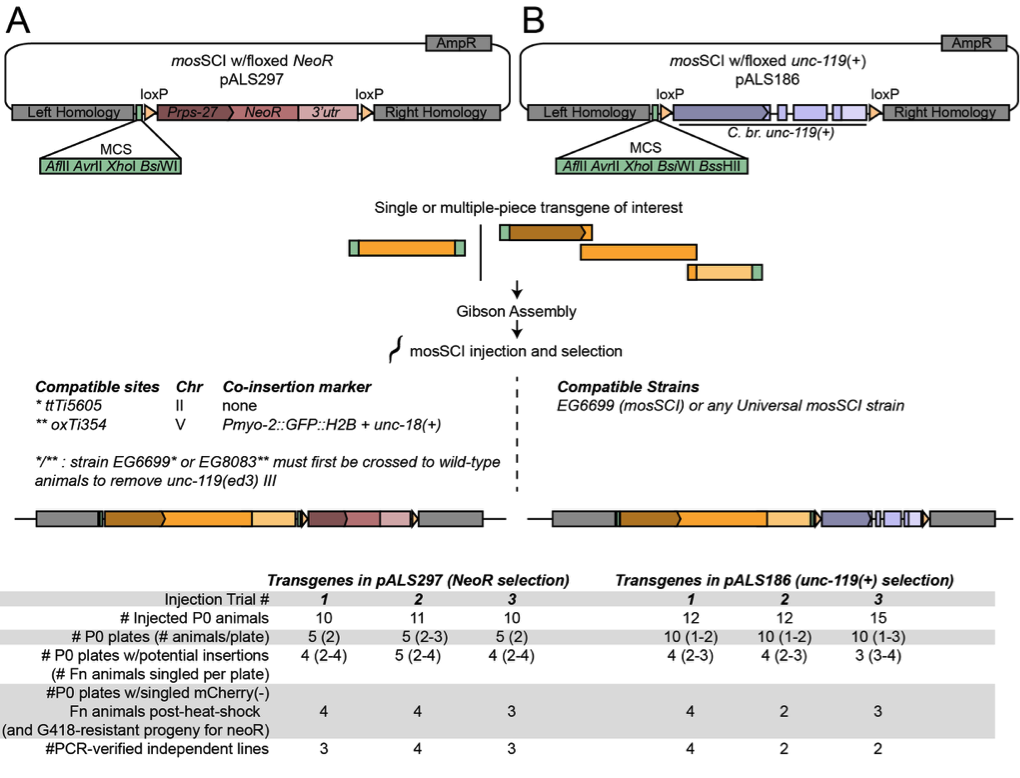
To add to the suite of tools for targeting ttTi5605, we constructed two modified mosSCI plasmids (Figure 1). First, we made a plasmid for ttTi5605-mosSCI using G418 selection. Antibiotic selection has been incorporated into the miniMos system for random transgene insertion (Frokjaer-Jensen et al. 2014), however existing ttTi5605-mosSCI plasmids use unc-119(+) selection. Our plasmid allows use of the ttTi5605 site without the need for injection into unc-119(ed3) mutants, which are more challenging to grow than wild-type animals and do not have optimal germline health on standard E. coli OP50 as a food source.
Second, we modified the standard unc-119 selection-based ttTi5605-mosSCI plasmid (pCFJ151) by adding loxP recombination sites flanking the unc-119(+) selection cassette (Figure 1B). This change allows excision of the unc-119(+) selection cassette using an existing Cre recombinase protocol (Dickinson et al. 2013). Since unc-119(+) is a frequently used selection marker, this new feature will add flexibility for construction of strains with multiple transgenes.
For increased versatility, both of the plasmids described here are minimal and do not encode additional tags or fluorescent proteins. Multiple-fragment transgenes can be seamlessly fused and cloned into these plasmids using the Gibson isothermal assembly technique by designing PCR amplicons, synthetic dsDNAs, restriction fragments or oligos with overlapping homologous ends (Gibson et al. 2009). We performed three independent test injections using different transgenes and found both of our plasmids to be effective for mosSCI (Figure 1). We also successfully excised the unc-119(+) cassette (Figure 1B) using Cre recombinase for two independent insertions (Methods). However, a caveat of the current NeoR plasmid (Figure 1A) is that no additional phenotypic marker was included between the loxP sites. Therefore, unlike unc-119(+) excision, which can be easily screened for using a visible phenotype, excision of NeoR would require screening by PCR or by loss of drug resistance. Overall, we hope that these excisable unc-119(+) and NeoR mosSCI plasmids will be useful companions to the extensive mosSCI plasmid and strain toolkit (Frokjaer-Jensen et al. 2008, Zeiser et al. 2011, Frokjaer-Jensen et al. 2012, Frokjaer-Jensen et al. 2014).
Reagents
Plasmid and Strain Construction. Plasmid pALS186 (Addgene # 129539; Table 1) was constructed in two steps from pCFJ151 (Addgene # 19330). Briefly, the C. briggsae unc-119(+) selection cassette was removed, leaving a modified minimal multiple cloning site (MCS) between the ttTi5605 insertion homology arms. A PCR amplicon containing C. br. unc-119(+) flanked by loxP sites was cloned into the BssHII(PteI)/BglII sites. To construct plasmid pALS297 (Addgene# 129538; Table 1), the loxP::C. br. unc-119(+)::loxP selection cassette was excised from pALS186 by BssHII(PteI)/BglII digestion and replaced by an MluI/BglII-digested PCR amplicon containing the NeoR selection cassette flanked by loxP sites. The NeoR selection cassette (Prps-27::NeoR::unc-54-3ʹutr) was amplified from pCFJ910 (Addgene # 44481).
To create strain ALS344, strain EG6699 (CGC; Table 1) was backcrossed 4X to wild-type N2 animals to remove the unc-119(ed3) allele. Segregation of unc-119(ed3) was followed by the unc phenotype. The ttTi5605(mos1) insertion site was followed using a three-oligo PCR (o508, o509, o91; Table 1) modified from the Wormbuilder site (http://www.wormbuilder.org/) to simultaneously amplify the ~720 bp product from the mos1 insertion allele and the 455 bp product from the wild-type allele. PCRs were performed with DreamTaq Green 2X PCR Mastermix (Thermo #K1081) using 1 uL of single worm lysate in a 20 uL reaction at an annealing temperature of 57°C.
Cloning transgenes into mosSCI plasmids. Transgenes of interest were assembled from multiple PCR amplicons with ~20 nucleotide overlapping ends into mosSCI plasmids by Gibson Isothermal assembly (Gibson et al. 2009) using a home-made enzyme mix or a kit (NEB #E2621S). For assembly into AvrII(XmaJI)/XhoI-digested pALS297 or pALS186, the first fragment of the transgene is amplified using the forward primer op297-p186AvrIIF (Table 1) and a transgene-specific reverse primer. The last fragment of the transgene is amplified using a transgene-specific forward primer and the reverse primer op297XhoIR or op186XhoIR (Table 1) for assembly into pALS297 or pALS186, respectively.
Microinjection and screening. Worm mosSCI transgenesis was performed according to the direct injection protocol (Frokjaer-Jensenet al., 2012) as described in detail on the Wormbuilder site (http://www.wormbuilder.org/). Transgene-containing mosSCI plasmids were purified by ZymoPURE plasmid miniprep (Zymo #D4209). Co-injected plasmids were purified by HiSpeed Plasmid Midi kit (Qiagen #12643). For unc-119(+)selection, plasmids were injected into strain EG6699 (Frokjaer-Jensen et al. 2012), which was grown on E. coli HB101. For NeoRselection, plasmids were injected into strain ALS344 (Table 1) and G418 was added to plates the day after injection as described (Frokjaer-Jensen et al. 2014) (125 uL of 25 mg/mL G418 per 35 mm plate).
Following heat-shock and counter-selection for co-injection markers, inserts were verified by PCR using pairs of primers where one primer hybridized to genomic DNA flanking the insertion site and outside of the vector homology arms (o145- 118 nt upstream of left homology arm, or o146- 63 nt downstream of right homology arm) and the second primer hybridized either within the transgene or selection cassette (Table 1). Amplification was carried out using either DreamTaq Green 2X PCR Mastermix (Thermo #K1081; for amplicons <2 kb) or Phusion polymerase (Thermo #F530L; for amplicons >2kb), respectively, according to the manufacturer’s instructions in 20 uL reactions containing 1 uL of a 6uL single worm lysate as the template.
Excision of the unc-119(+) selection cassette. The C. br. unc-119(+) selection cassette was excised using Cre recombinase as described (Dickinson et al., 2013) using pDD104 (Peft-3::Cre; Addgene # 47551). Briefly, 15-20 animals were injected and moved onto 5-6 plates (3 P0 worms/plate). Approximately 15 F1 animals expressing the mCherry co-injection marker were singled. From the progeny of these animals, unc F2 animals were singled to establish excised lines. At least two independent lines were recovered from each of two independent trials. Cre-mediated excision of the NeoR selection cassette has not been tested.
| Plasmids |
| pALS297 (ttTi5605-MCS loxP::Prps-27::NeoR::unc-54-3ʹutr::loxP); Addgene # 129538 |
| pALS186 (ttTi5605-MCS loxP::C.br.unc-119(+)::loxP); Addgene # 129539 |
| Strains |
| EG6699 ttTi5605(mos1) II; unc-119(ed3), CGC, note lost extrachromosomal array oxEx1578 |
| ALS344 ttTi5605(mos1) II, this study; EG6699 was crossed 4x to N2 to remove the unc-119(ed3) allele |
| Primers to follow ttTi5605(mos1) II |
| o508-mos1F(oJL103) TCTGCGAGTTGTTTTTGCGTTTGAG |
| o509-ttTi5605F GATTGTTTGACCTGGCGGAACT |
| o91-ttTi55605R(NM3888) ACGCCCAGGAGAACACGTTAG |
| Primers for cloning |
| op297-p186AvrIIF TAGAGGGTACCAGAGCTCACCTAGGN*20-22 |
| op297XhoIR GTTATaCGCGCACCGTACGTCTCGAGN*20-22 |
| op186XhoIR GTTATgCGCGCACCGTACGTCTCGAGN*20-22 |
| Primers for insert verification |
| 5ʹarm, neoR mosSCI |
| o145-ttTi5605outerF AGGCAGAATGTGAACAAGACTCGAG (NM3880 + 2nt) |
| o496-unc54-3ʹutrR GGCCCAGACGTGCGAAGAAATA
(or transgene-specific REV primer for long transgenes) |
| 3ʹarm, unc-119 mosSCI |
| o145-ttTi5605outerF AGGCAGAATGTGAACAAGACTCGAG (NM3880 + 2nt) |
| o147-Cbr-unc-119promR AAGTAGCAGAGCTGGGGAGAAGAA
(or transgene-specific REV primer for long transgenes) |
| 5ʹarm, neoR mosSCI |
| o311-NeoRF CTTCTATCGCCTTCTTGACGAG |
| o146-ttTi5605outerR AATCGGGAGGCGAACCTAACTGT (NM3884 + 2nt) |
| 3ʹarm, unc-119 mosSCI |
| Transgene-specific FOR primer |
| o146-ttTi5605outerR AATCGGGAGGCGAACCTAACTGT (NM3884 + 2nt) |
Table 1: List of plasmids, strains and oligonucleotides. Oligonucleotides with NM or JL designations were described by or adapted from Dr. M. Nonet (Washington University School of Medicine in St. Louis) or Dr. J. L. Bessereau (Universitéde Lyon), respectively, based on protocols referenced on the Wormbuilder site (http://www.wormbuilder.org/). *, N20-22 represents 20-22 transgene-specific nucleotides.
Acknowledgments
We thank J.A. Calarco (University of Toronto) for assistance with microinjections. We thank Dr. Erik Jorgensen for pCFJ151 (ttTi5605_MCS; Addgene plasmid # 19330) and Dr. Bob Goldstein for pDD104 (Peft-3::Cre; Addgene plasmid # 47551). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
Funding
This work was supported by the University of Toronto (Faculty of Arts and Science, Dept. of Cell and Systems Biology) and the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant RGPIN-2019-06843).
Reviewed By
Christian Frøkjær-JensenHistory
Received: July 31, 2019Accepted: August 6, 2019
Published: August 27, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Aram, R; MacGillivray, K; Li, C; Saltzman, AL (2019). Tools for Mos1-mediated single copy insertion (mosSCI) with excisable unc-119(+) or NeoR (G418) selection cassettes. microPublication Biology. 10.17912/micropub.biology.000146.Download: RIS BibTeX