Corrections
Corrections to this article were made. These corrections were recorded in an Erratum published on Aug 23, 2019.
Description
Behavioral quiescence during sleep states can be measured by a variety of methods, each with advantages and limitations (Nagy et al., 2014). Centroid tracking gives information about locomotion (place-to-place movement) but may not register small head movements or other changes in body posture that do not change centroid position. Frame subtraction methods, based on analysis of pixel intensity changes between successive frames, register any movement as non-quiescent, but do not categorize the nature of movement. Counting body bends is a close proxy to locomotion (Karbowski et al., 2006) but can be time-consuming if done manually. Last, while nose tip movement has been observed to approximate body movement during C. elegans sleep (Bringmann, 2011), this approximation may not apply to mutants defective in only one of these behaviors. Here we provide an example of such a case.
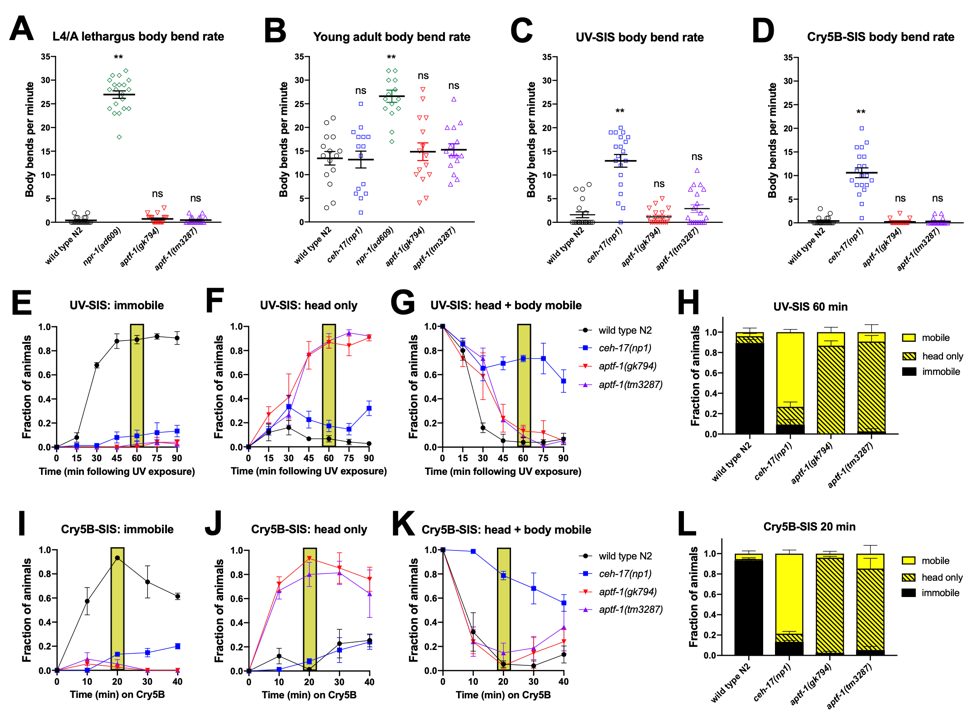
C. elegans undergoes robust developmentally-timed sleep (DTS), also known as lethargus, prior to each larval molt (Raizen et al., 2008). RIS interneuron-defective aptf-1(loss-of-function) mutants have been found to be impaired for movement quiescence during DTS based on tracking the speed of the worm’s nose tip (Turek et al., 2013). To determine whether this nose movement reflects locomotor activity, we quantified the locomotor rate of these animals, while blinded to genotype, by counting the number of sinusoidal body bends per minute. We compared lethargus (A) and post-lethargus (B) movement of two aptf-1(lf) strains to wild type (N2) and to lethargus-defective npr-1(lf) animals (Choi et al., 2013). We found that unlike npr-1(lf), which showed a high frequency of body bends during and outside of lethargus, aptf-1(lf) mutants greatly reduced their frequency of body bends during lethargus to a level similar to that of N2 controls. However, we noted that these RIS-defective animals were strikingly defective in head movement quiescence and also showed rocking movements: alternating backward and forward body movements, each resulting in less than a half-body translation of the worm’s position and virtually no net movement in either direction.
In addition to developmental sleep, C. elegans engages in sleep following exposure to conditions that cause cellular stress (Hill et al., 2014; Nelson et al., 2014; DeBardeleben et al., 2017). This stress-induced sleep (SIS) is dependent on the peptidergic ALA interneuron and the action of several neuropeptides that collectively promote a state of coordinated behavioral quiescence (Nath et al., 2016). Because the RIS neuron has recently been implicated in regulating SIS based on frame subtraction methods (Grubbs et al., 2019; Konietzka et al., 2019), we examined the behavior of aptf-1(lf) animals during SIS triggered by ultraviolet light (C) and by ingestion of pore-forming Cry5B toxin (D). We compared the behavior of aptf-1(lf) mutants to N2 and to SIS-defective ceh-17(lf) animals that are impaired for ALA neuron function (Van Buskirk and Sternberg, 2010). In contrast to ALA-defective animals, aptf-1(lf) mutants showed wild type body-bend quiescence during SIS. As in lethargus, RIS-defective animals showed head movement, but they did not rock back and forth as they did during lethargus. In an independent assay, we examined each animal (again, blinded to genotype) for five seconds every 10-15 min following exposure to a SIS trigger and categorized their movement during that time as “fully immobile”, “body immobile but head mobile”, where head mobility was defined as any discernible movement, or “head and body mobile”, where body mobility was defined as a translation of the body position by at least 1/10 body length. Most aptf-1(lf) animals moved only their heads and did not translate their body position (F and J); SIS body movement quiescence in aptf-1(lf) was similar to wild type, but took longer to set in in the case of UV-SIS (G). These data indicate that the RIS neuron plays a major role in controlling head movement quiescence, with a relatively minor impact on body movement quiescence, at least when examined on standard NGM plates. It will be of interest to determine whether the impact on body movement quiescence is a secondary consequence of head movement. Though head movement is often referred to as foraging behavior, we did not observe any feeding (pharyngeal pumping) in aptf-1 mutants in these assays. Our results have implications regarding the circuits regulated (head-only versus head and body) by RIS and ALA activity. This analysis also emphasizes the importance of visual inspection, scored blind to genotype, as a valuable tool in studies of quiescence.
Reagents
Strains available from the CGC: N2 wild type, IB16 ceh-17(np1), DA609 npr-1(ad609), HBR227 aptf-1(gk794), HBR232 aptf-1(tm3287). Cry5B-expressing bacteria, received from Raffi Aroian, is available from our lab.
Acknowledgments
Many thanks to David Raizen for helpful comments on this microPublication, and to the worms for reminding us that we need to look at them.
References
Funding
This work was supported by a National Science Foundation Faculty Early Career Development Program (CAREER) award IOS#1553673 to C.V.B. Strains were kindly provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
Ann Hart and Kelsey SchuchHistory
Received: August 1, 2019Accepted: August 8, 2019
Published: August 19, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Robinson, B; Goetting, DL; Cisneros Desir, J; Van Buskirk, C (2019). aptf-1 mutants are primarily defective in head movement quiescence during C. elegans sleep. microPublication Biology. 10.17912/micropub.biology.000148. Erratum in: microPublication Biology. 10.17912/micropub.biology.000162.Download: RIS BibTeX