Description
Xenopus laevis is an excellent model organism for studies on vertebrate endoderm development (Womble et al., 2016; Zorn, 2009). The endoderm contributes to a number of important organs including the lungs, liver, gall bladder, pancreas, stomach and intestine. Each of these organs differentiate from the foregut, midgut, and hindgut domains of the early endoderm (Chalmers and Slack, 2000). It has recently been shown that as early as gastrulation differential fates are being specified within the endoderm (Costa et al., 2003; Rankin et al., 2018). Several studies have worked to identify markers of different fates in the developing endoderm in order to assist with research on endoderm specification, differentiation, and organogenesis, but not all of the identified genes have known functions and some are currently unclassified (Chen et al., 2003; Costa et al., 2003; Park et al., 2007; Zorn and Mason 2001).
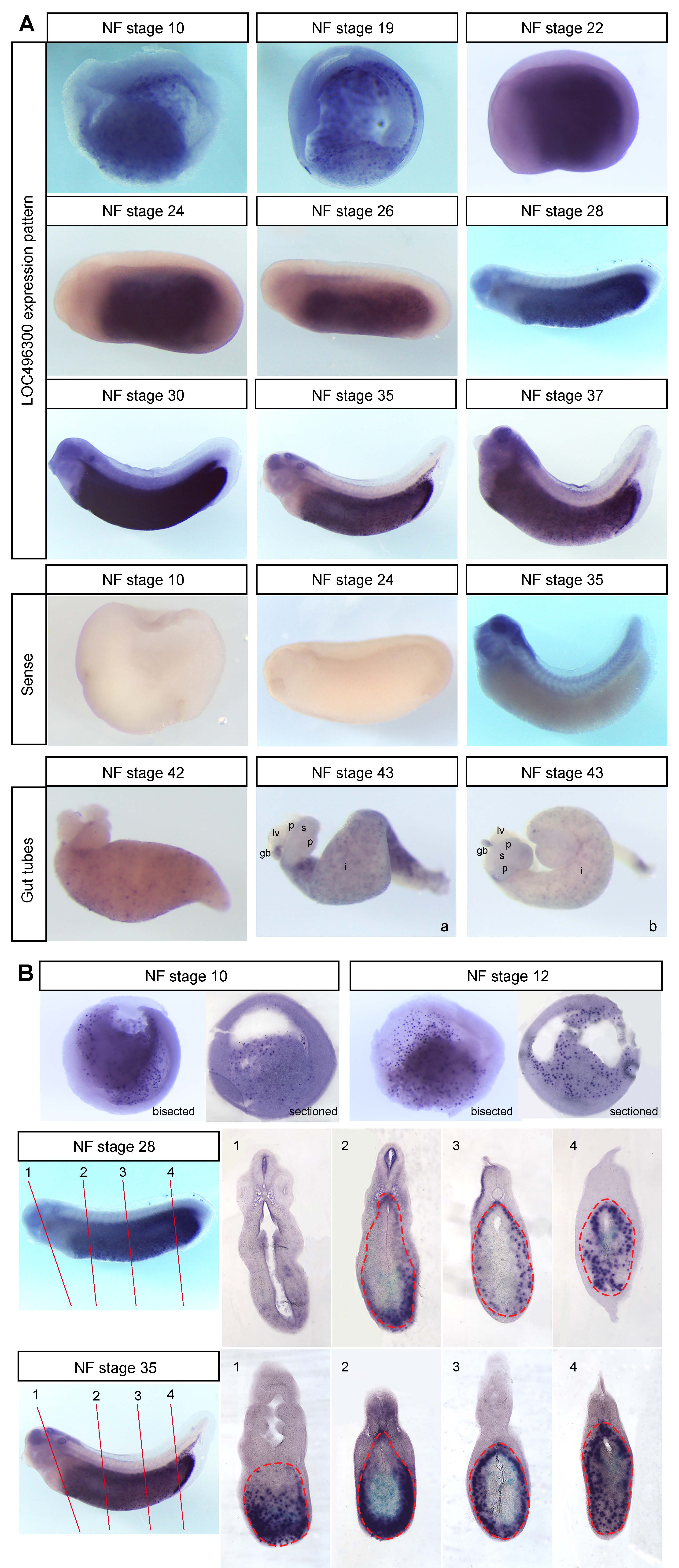
We performed whole mount in situ hybridization (WISH) on hypothetical locus LOC496300 (Xl.8755; Ref Seq NM_001095458.1) and found that it is expressed throughout the endoderm of early Xenopus embryos (NF stage 10-23) and as development proceeds it becomes localized to the midgut and hindgut endoderm (NF stage 27-37) (Figure 1A), which later gives rise to the intestine posterior to the stomach (Chalmers and Slack, 1998). LOC496300 has a similar expression pattern to other midgut-hindgut specific markers like ctbs, impdh1, and darmin during the early tadpole stages (Costa et al., 2003; Pera et al., 2003). LOC496300 does show some expression in the foregut early on, whereas darmin is excluded from the foregut even at gastrulation (Costa et al., 2003, Pera et al., 2003). We repeated the WISH for LOC496300 with three sets of embryos and saw consistent expression patterns with each set (examining a total of at least 18 embryos for each stage). WISH with the sense probe for LOC496300 as a control showed no staining in the endoderm at any stage, but non-specific staining in the brain, eye, and pharynx at NF stage 35 (see Figure 1A).
Sectioned embryos show expression of LOC496300 in the endoderm is punctate in a subset of cells throughout all regions of the early endoderm (Figure 1B). At tailbud stages LOC496300 is expressed in both the dorsal and ventral midgut and hindgut and in the gut tube at NF stage 42 and 43, maintaining a punctate expression pattern throughout the intestine. It is not clear what subset of cells might be expressing LOC496300. There are several other genes that show punctate expression patterns in the endoderm such as Sox17α, Sox17β, and darmin at NF stage 10 when there are many yolk granules still present in the endoderm cells (Costa et al., 2003; Hudson et al., 1997; Sinner et al., 2004). Later in development, it is possible LOC496300 is expressed in a group of cells that will differentiate into a specific cell type for the functional intestinal epithelium of the feeding tadpole, which consists mainly of principal absorptive cells as well as some gland cells, endocrine cells and lymphocytes (Marshall and Dixon; 1978; Shi and Ishizuya-Oka, 1996). This larval epithelium will eventually undergo apoptosis except for a few cells that de-differentiate into progenitor stem cells which give rise to the adult intestinal epithelium at metamorphosis (Ishizuya-Oka, 1996; Ishizuya-Oka, 2011; Ishizuya-Oka et al., 2009; Shi and Ishizuya-Oka, 1996; Sun et al., 2013). It would be interesting to determine which genes expressed in the embryonic intestine, like LOC496300, play a role in larval intestinal development and function, and for the dramatic transition that occurs during metamorphosis.
LOC496300 is currently unclassified. Searching for conserved domains identified a Spc7 kinetochore domain in LOC496300, suggesting it may play a role in kinetochore function. It also has KOG2044 and SbcC exonuclease domains, suggesting a possible role in replication, recombination, or repair. Further research into the functions of LOC496300 will help identify the role it plays specifically in the endoderm during development and whether it contributes to the specification and differentiation of the intestinal fate. Additionally, it would be interesting to discover whether LOC496300 is regulated by dynamic Wnt, BMP, and RA signals during different stages of development, as others have shown that coordination of these signaling pathways specifies different endodermal fates (Rankin et al., 2018; Stevens et al., 2017).
Reagents
Xenopus laevis embryos were cultured and gut tubes were isolated using standard protocols approved by the NKU Institutional Animal Care and Use Committee (IACUC Protocol #2014-09). The plasmid to make the WISH probe for LOC496300 was purchased sequence-verified from Source Bioscience (CF286219 clone #IRBHp990A1273D) and we used SalI to linearize the plasmid and T7 to transcribe the anti-sense RNA probe (NotI with SP6 for the sense probe). We followed previously described protocols for WISH (Costa et al., 2003). For sectioning, the embryos were embedded in gelatin-albumin and cut into 50uM sections using a Leica vibratome VT1200 S.
Acknowledgments
Thank you to all the members of the Shifley lab for their assistance.
References
Funding
This work was supported by the Kentucky Biomedical Research Infrastructure Network NIGMS grant #8P20GM103436 as well as the Northern Kentucky University Research Foundation, Northern Kentucky University Faculty Senate, and NKU Center for Integrative Natural Science and Mathematics (CINSAM).
Reviewed By
Sang-Wook Cha and Christina James-ZornHistory
Received: June 26, 2019Accepted: August 2, 2019
Published: August 12, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Stewart, ME; Donahue, KM; Wilke, EG; Shifley, ET (2019). LOC496300 is expressed in the endoderm of developing Xenopus laevis embryos. microPublication Biology. 10.17912/micropub.biology.000150.Download: RIS BibTeX