Department of Biology, Hopkins Marine Station of Stanford University, Pacific Grove, CA 93950
Description
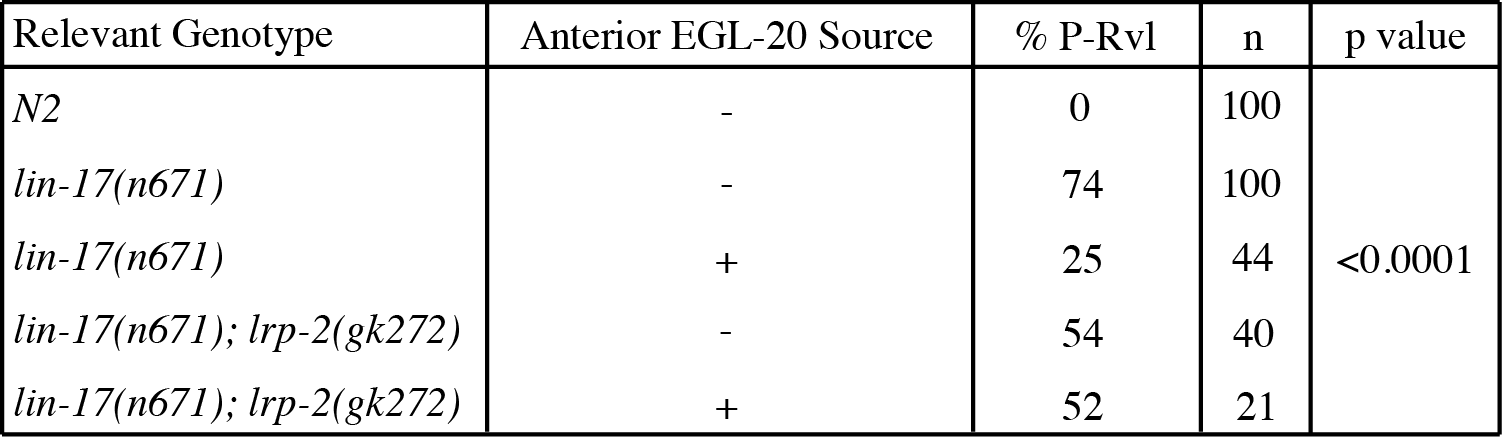
The C. elegans vulva is formed from divisions of three vulval precursor cells (VPCs) – P5.p, P6.p, and P7.p – arranged along the anteroposterior axis in the ventral epithelium (Sulston and Horvitz, 1977). Previous analyses show the orientation of P5.p and P7.p descendants is determined by the interaction of multiple Wnt signals. egl-20/Wnt is expressed in the tail (Whangbo and Kenyon, 2000) and forms a posterior-to-anterior concentration gradient (Coudreuse et al., 2006). It has previously been shown that EGL-20 acts instructively during vulva development by imparting directional information, as opposed to being permissive, where it would only be required for polarization (Green et al., 2008; Minor et al., 2013). By moving the source of egl-20 expression from the posterior of the worm to the anchor cell, the axis of symmetry of the developing vulva, we can reorient the daughter cells of P5.p and P7.p toward the center in a wild-type configuration.
Expression of egl-20 from the center of the axis of symmetry partially suppresses the lin-17(n671) phenotype (Green et al., 2008; Table 1). To test whether LRP-2 acts downstream of EGL-20, we ectopically expressed egl-20 from the anchor cell in a lin-17(n671); lrp-2(gk272) double mutant background and compared it to a lin-17(n671); lrp-2(+) strain. If LRP-2 acts downstream of EGL-20, then anteriorly-expressed EGL-20 will not be able to suppress the lin-17 phenotype, with is the result observed (Table 1). Thus, like CAM-1 and VANG-1, LRP-2 likely acts downstream of EGL-20.
Reagents
Strains:
VC543: lrp-2(gk272). Strain obtained from the CGC and provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which is part of the international C. elegans Gene Knockout Consortium.
MT1306: lin-17(n671) (Ferguson and Horvitz, 1985)
PS5800: lin-17(n671); syEx1031[Pfos-1a::EGL-20::GFP] (Green et al., 2008)
MT1488: lin-17(n671); unc-13(e1091)
The lin-17(n671); lrp-2(gk272) double mutant constructed by crossing lrp-2(gk272) males with strain MT1488 hermaphrodites.
References
Funding
Howard Hughes Medical Institute, with whom PWS was an Investigator. The National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number 1F32NS098658-01A1 awarded to PJM.
Reviewed By
David EisenmannHistory
Received: July 29, 2019Accepted: August 26, 2019
Published: August 27, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Minor, PJ; Sternberg, PW (2019). LRP-2 likely acts downstream of EGL-20/Wnt. microPublication Biology. 10.17912/micropub.biology.000153.Download: RIS BibTeX