Department of Biology, Hopkins Marine Station of Stanford University, Pacific Grove, CA 93950
Description
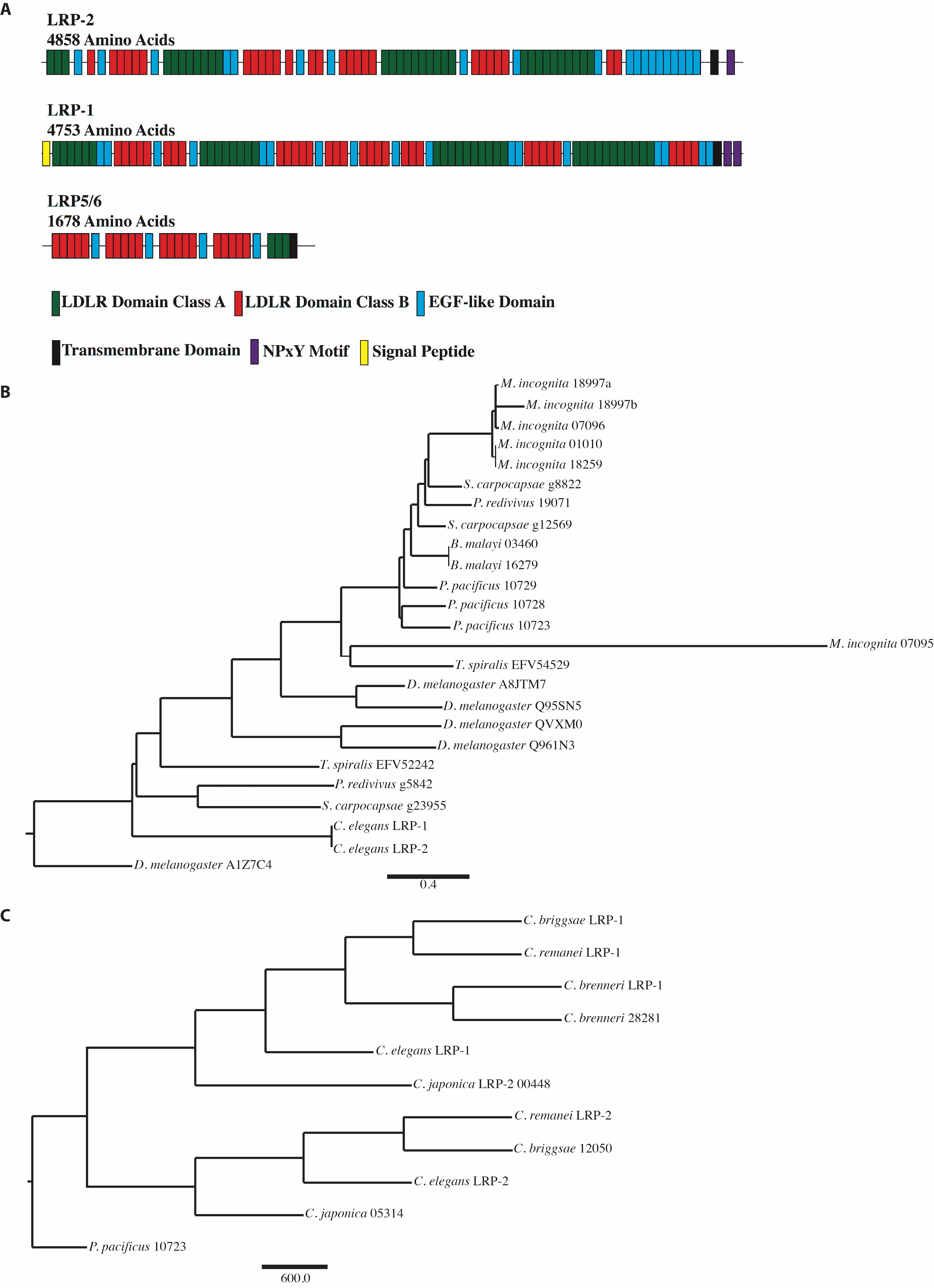
The regulation of vulval cell lineage polarity is controlled by Wnt signaling. Previously known components involved in the regulation of vulval cell lineage polarity include LIN-17, LIN-18, CAM-1, and VANG-1 (Inoue et al., 2004; Gleason et al., 2006; Green et al., 2008). A directed bioinformatics screen of known Wnt pathway components was performed to find additional genes involved in directing vulval orientation. A BLAST was run using other known Wnt receptors and it was determined that C. elegans does not contain a true ortholog of Drosophila LRP5/6 (Arrow) (He et al., 2004; Eisenmann, 2005), but does have multiple low-density lipoprotein receptors, including LRP-1 and LRP-2 (Figure 1). Like other low-density lipoprotein receptors, both LRP-1 and LRP-2 contain many LDLR Domain Class A and Class B repeats, EGF-like domains, and a transmembrane domain. However, having approximately three times as many amino acids, LRP-1 and LRP-2 are more similar to megalin than LRP5/6 (Yochem et al., 1999). The absence of LRP5/6 within C. elegans but presence in flies and all other higher order organisms suggests that the gene encoding LRP5/6 arose after nematodes, potentially from either LRP1 or LRP2/megalin, as both receptors contain the entire extracellular portion of LRP5/6 in a single contiguous sequence block (Figure 1).
Our examination of the protein sequence of LRP-1 and LRP-2 indicates that most nematodes have at least two copies of LRP-like proteins with C. elegans LRP-1 and LRP-2 being highly similar possibly due to a recent duplication and divergence (Figure 2). Comparing the sequences across Caenorhabditis we find that LRP-1 proteins cluster together and LRP-2 proteins also form their own cluster. Based on location in the genome and sequence similarity from protein alignment, we believe that Caenorhabditis lrp-2 is a recent duplication and divergence of lrp-1 (Figure 2).
Methods
Request a detailed protocolAvailable predicted protein datasets from nematodes were obtained from WormBase release WS225 (www.wormbase.org). Other sequences were obtained from the NCBI/NIH repository (ftp://ftp.ncbi.nih.gov/genomes). Maximum likelihood (ML) analyses with 1,000 bootstraps were done using the RAxML BlackBox server (http://phylobench.vital-it.ch/raxml-bb). Protein domain analysis performed using the SMART protein domain analysis website (http://smart.embl-heidelberg.de)
References
Funding
Howard Hughes Medical Institute, with whom PWS was an Investigator. The National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number 1F32NS098658-01A1 awarded to PJM.
Reviewed By
David EisenmannHistory
Received: July 29, 2019Accepted: August 26, 2019
Published: August 27, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Minor, PJ; Sternberg, PW (2019). Low density lipoprotein receptors LRP-1 and LRP-2 in C. elegans. microPublication Biology. 10.17912/micropub.biology.000154.Download: RIS BibTeX