School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW 2006, Australia
Description
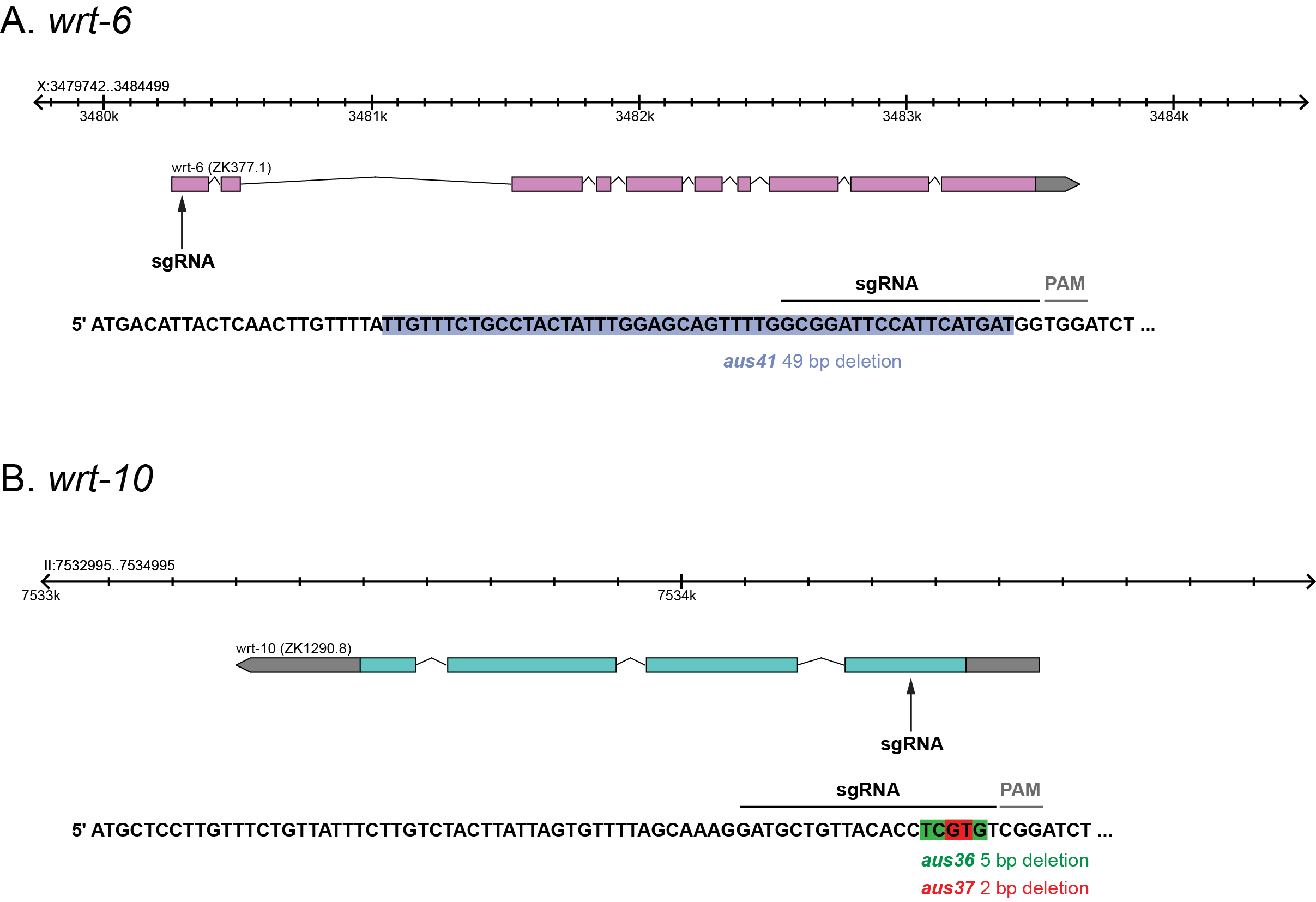
We have generated new putative null alleles of two members of the C. elegans Hedgehog-related pathway: wrt-6 and wrt-10. The warthog (wrt) family member wrt-6 encodes a predicted secreted signalling molecule that contains an N-terminal Wart domain and a C-terminal autoprocessing domain (Hint or Hog domain) (Bürglin 1996; Aspöck et al. 1999; Bürglin, 2008). wrt-10 encodes another member of this Wrt family that contains the Wart domain but lacks the Hog domain (Bürglin 1996; Aspöck et al. 1999).
To generate deletion alleles, we used CRISPR-Cas9 to target the first exon of wrt-6 or wrt-10 (wrt-6 sgRNA: 5′-GATGCTGTTACACCTCGTGT-3′, wrt-10 sgRNA: 5′-GCGGATTCCATTCATGATGG-3′). We isolated the wrt-6 (aus41) allele that contains a 49 bp out-of-frame deletion, predicted to cause a premature stop codon after 7 amino acids (Figure 1A). We also generated two wrt-10 deletion alleles: aus36 and aus37. The aus36 allele contains a 5 bp deletion, predicted to cause a premature stop codon after 33 amino acids (Figure 1B). The aus37 allele contains a 2 bp deletion, predicted to cause a premature stop codon after 34 amino acids (Figure 1B). As all these alleles are predicted to cause premature stop codons in the first exon of each gene, they are likely to represent molecular nulls. We found that these alleles are viable and have no obvious gross morphological phenotype. Therefore, in-depth phenotypic examination is required to dissect their functional role. To conclude, we have isolated specific deletion alleles for wrt-6 and wrt-10 and will therefore help reveal new insights into the functions of Hedgehog-related signaling in C. elegans.
Methods
Request a detailed protocolsgRNA target sequences were incorporated into a pU6::klp-12 sgRNA expression vector by PCR (Norris et al. 2015). Wild-type (N2) animals were injected with a mix consisting of the sgRNA expression vector (250 ng/µL), Cas9 expression vector (peft-3::cas9::tbb-2) (50 ng/µL) (gift from the de Bono Lab), pCFJ90 (pmyo-2::mCherry::unc-54) (2.5 ng/µL) and pCFJ104 (pmyo-3::mCherry::unc-54) (5 ng/µL). We used a 3-primer method for screening for deletions in mCherry-positive animals (external forward, internal forward directly upstream of PAM site, and external reverse; see Reagents for sequences). The external forward and reverse primers were used for Sanger sequencing.
Reagents
Oligonucleotides used to detect and genotype wrt-6(aus41)
Expected PCR amplicon: Wild-type: 578 and 327 bp. Mutant: 529 bp
External forward: GTGTCGTCGGATTTCTTCATTC
Internal forward: GCGGATTCCATTCATGATGG
External reverse: GAGCCACTTACAGTCCAAGG
Oligonucleotides used to detect and genotype wrt-10(aus36) and (aus37)
Expected PCR amplicon: Wild-type: 539 and 241 bp. Mutant: 534 bp (aus36), 537 bp (aus37)
External forward: GCAACAATCGGTCCGTCCAC
Internal forward: GATGCTGTTACACCTCGTGT
External reverse: GGTTTCAGCACGGCAAGAAG
Strains
HRN666 wrt-10(aus36) II
HRN667 wrt-10(aus37) II
HRN680 wrt-6(aus41) X
Strains have not been outcrossed. Will be available at the CGC.
References
Funding
This work was supported by an Australian Government Research Training Program (RTP) Scholarship to T.S, and the National Health and Medical Research Council (GNT1137645 to R.P).
Reviewed By
AnonymousHistory
Received: September 11, 2019Accepted: October 8, 2019
Published: October 15, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Sherry, T; Nicholas, HR; Pocock, R (2019). New deletion alleles for Caenorhabditis elegans Hedgehog pathway-related genes wrt-6 and wrt-10. microPublication Biology. 10.17912/micropub.biology.000169.Download: RIS BibTeX