Department of Cell Biology, Rowan University School of Osteopathic Medicine, Stratford, NJ 08084, USA
Description
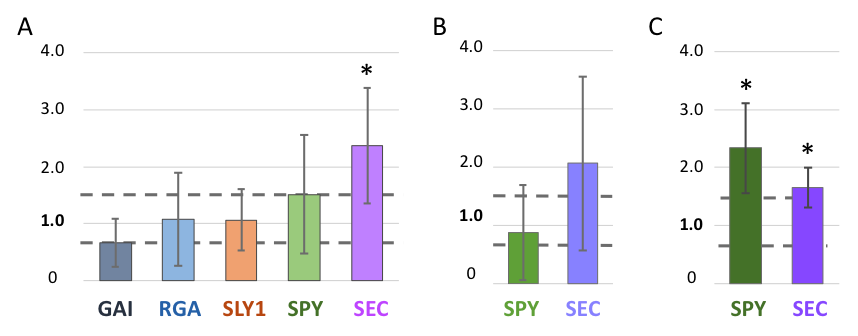
Since trichome number is altered with disruption of GCN5 or ADA2b (Kotak et al. 2019; Wang et al. 2019), we sought to determine whether these chromatin factors might impact the expression of known regulators of trichome initiation, including two members of the DELLA family of repressor proteins, GA-Insensitive (GAI) and REPRESSOR of ga1-3 (RGA). As their names suggest, these proteins are sensitive to gibberellin (GA) signaling (Silverstone et al. 2001). GA complexed with its receptor can bind to DELLA proteins and promote their interaction with a component of an E3 ubiquitin ligase, SLEEPY1 (SLY1), leading to subsequent proteasome-media degradation (Dill et al. 2004). In addition to SLY1, we also assessed the expression of SPINDLY (SPY) and SECRET AGENT (SEC), both known to covalently modify RGA and impact its functionality in a positive and negative manner respectively (Zentella et al. 2016, 2017).
In mature rosette leaves of gcn5-6 plants (harboring a T-DNA insertion that disrupts the catalytic domain of GCN5; Kotak et al. 2018), we detect a trend of decreased expression of GAI, while no consistent change in expression is seen for RGA. Among the factors that influence the activity of the DELLA repressors, SLY1 showed no change in expression. On average, there was a 1.5X increase in SPY expression in gcn5-6 mutants while SEC expression was more than doubled (Fig. 1A). Expression analysis comparing the Columbia (wildtype) and gcn5-6 background included five biological replicates, except for SLY1, where n=4.
A microarray experiment conducted by Vlachonasios et al. 2003 provides some additional data on these genes’ expression in ADA2b and GCN5 disruption mutants. Specifically, in ada2b-1 and gcn5-1 plants (Vlachonasios et al. 2003), GAI expression is slightly decreased (less than three-fold) while RGA expression is unchanged, paralleling what we report here with gcn5-6. SLY1 expression was also slightly decreased in ada2b-1 and gcn5-1 plants. Since SPY and SEC were not included in the microarray experiment, we assayed their expression via qRT-PCR. SPY expression is unchanged in gcn5-1. This is consistent with the lack of a significant change in gcn5-6 plants. In ada2b-1 plants, SPY expression is increased more than 2-fold. SEC expression is increased in both gcn5-1 and ada2b-1 in comparison to Ws (wildtype) controls. These experiments included 4-6 biological replicates.
Reagents
Arabidopsis thaliana plants were germinated in soil and grown at 22°C under continuous light conditions (140 µmoles/m2/sec). Plants were watered once a week with Hoagland’s solution and a second time weekly (or as needed) with deionized water. Plants were genotyped via PCR as described previously (Kotak et al. 2018) and rosette leaves were harvested at 4-5 weeks of age and flash frozen in liquid nitrogen prior to preparation of RNA (Qiagen Plant RNeasy kit). Between 0.5-1 µg of RNA was used in each cDNA synthesis reaction (Maxima H Minus cDNA Synthesis Master Mix with dsDNAse, Thermo Scientific). Quantitative PCR was performed using an Applied Biosystems StepOne Plus instrument with the FAST SYBR Green Master Mix. Primer sequences used to amplify each target cDNA as well as two “housekeeping” genes, At4g26410 (used in GAI and RGA trials) and At1g13440 (used in SLY1, SPY, and SEC trials) are listed in the Table below. Comparative Ct analysis that includes normalization of wildtype (Ws-2 or Col-0) and mutant samples based on expression of the housekeeping gene was conducted using the Applied Biosystems instrument software. Melting temperature analysis confirmed the presence of a single amplicon in each reaction. Each data point within an experiment was performed in triplicate, with an allowable standard deviation of Ct values < 5%. Average Ct values for the housekeeping gene for wildtype and mutant samples needed to be within three cycles of one another for data to be included in this analysis.
| Gene name | Locus identifier | Forward Primer | Reverse Primer |
| GAI | AT1G14920 | CTGTGGTTGAGCAGGAATCG | AACCTCCGACATGACCTTGT |
| RGA | AT2G01570 | GAAAGTTCTCGGCGTTGTGA | CTTGCTCAACCACCGTGAAA |
| SLY1 | AT4G24210 | TGCTCACAACGAGACAAACA | TGCTCACGCAAGATGACATA |
| SPY | AT3G11540 | TTGAGGCTCACAGAGACTGG | GGTGATAGGTCGCTCTGGAT |
| SEC | AT3G04240 | TCTGCTATGTCATCTGATGCTATCG | GGAAGCCCATATATGAAACCTGGAT |
| RHIP1 | AT4G26410 | GAGCTGAAGTGGCTTCCATGAC | GGTCCGACATACCCATGATCC |
| GAPC2 | AT1G13440 | TTGGTGACAACAGGTCAAGCA | AAACTTGTCGCTCAATGCAATC |
Acknowledgments
We would like to thank Dr. Konstantinos Vlachonasios (University of Thessaloniki, Greece) for his helpful comments on this manuscript and for his on-going collaboration. We also thank members of the BIO 220-11 class, Spring 2019 at Muhlenberg College for contributions to the SPY/gcn5-6 data.
References
Funding
We thank Muhlenberg College for support of this research.
Reviewed By
Neil OlszewskiHistory
Received: September 12, 2019Accepted: October 7, 2019
Published: October 17, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Trachtman, N; Sockler, P; Caiola, H; McCain, ER; Hark, AT (2019). Expression of the DELLA repressor GAI and its regulators SPY and SEC are impacted by disruption of chromatin modifiers. microPublication Biology. 10.17912/micropub.biology.000175.Download: RIS BibTeX