present address: Department of Cell and Developmental Biology, University College London
Department of Biological Sciences, HHMI, Columbia University
Description
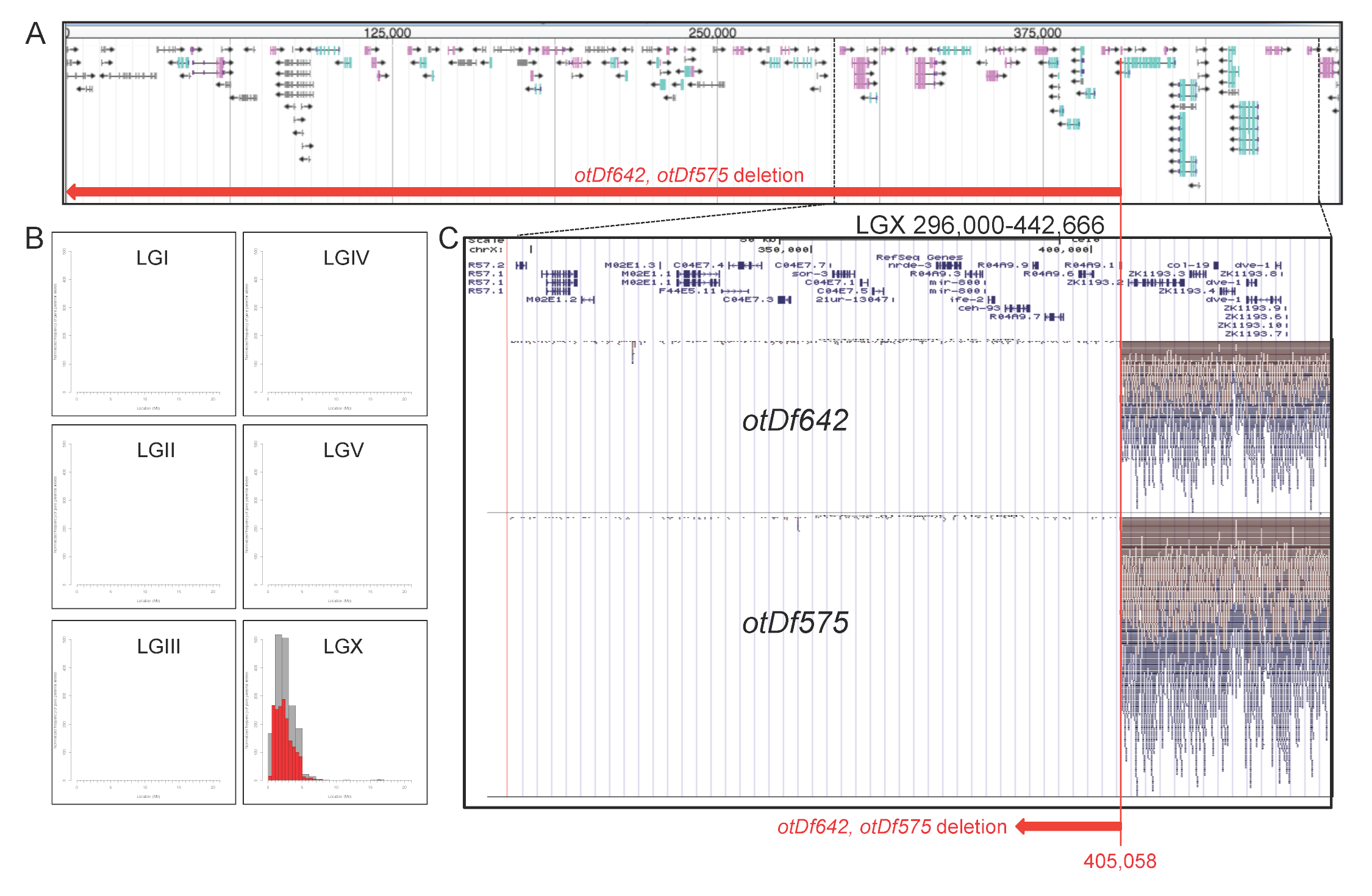
We are interested in isolating mutants that affect lineage specification in the nervous system of the nematode C. elegans. The harsh touch sensory neuron PVD, generated by the postdeirid lineage, can be labeled with two transgenically expressed fluorophores, ser-2::gfp (otIs138 transgene)(Tsalik et al., 2003) and dop-3:rfp (vsIs33 transgene)(Chase et al., 2004). Using a otIs138; vsIs33 double transgenic strain, we screened for EMS-induced mutations in which both markers fail to be expressed in PVD and isolated two strains in which the majority of animals fail to display reporter expression in PVD (ot642 mutant strain: 56% animals showed no marker expression in PVD expression; ot575 mutant strain: 71% of animals; n=54). ot575 and ot642 fail to complement each other. Animals that carry these alleles are viable, fertile and display no obvious morphological abnormalities. Both strains were subjected to Illumina whole genome re-sequencing, one in combination with Hawaiian SNP mapping (ot642)(Doitsidou et al., 2010) the other in combination with variant discovery mapping (ot575)(Minevich et al., 2012). These two orthogonal mapping approaches revealed that both mutant strains carry the exact same alteration: a loss of 405,058 bp from the extreme left end of the X chromosome (Figure 1). ot575 and ot642 were isolated from separate rounds of screens so these mutants were not progeny of the same initial parent. Each of these strains were isolated with the otIs138[ser2prom3::gfp] transgene that is also located on chromosome X so it is possible that this transgene somehow contributes to chromosome instability in some way or form.
We confirmed the deletion with PCR primers located within this deletion, which yielded a PCR product from wild-type animals, but not mutant animals. Since the size of the deletions classifies these alleles as deficiencies, we renamed these alleles otDf575 and otDf642.
The deletion in otDf575 and otDf642 eliminates 50 protein-coding genes (one of them, R04A9.1, which carries the deletion breakpoint, is cut in half), at least 20 pseudogenes and a number of regulatory RNAs, ranging from several 21U RNAs to three miRNA encoding genes, mir-258.1, mir-258.2 and mir-800. The protein coding genes lost in these strains include a number of different functional categories, including a number of gene regulatory factors: two transcription factors (elk-2 and ceh-93, a homeobox gene), two SET-domain containing chromatin factors (set-28, set-33), an argonaute protein (nrde-3) and the polycomb-group gene, sor-3.
We find that the sor-3 locus alone is responsible for the PVD mutant phenotype of otDf575 animals, because otDf575 fails to complement sor-3(bp185) (5/21 ot575/bp185 transheterozygous animals show PVD loss) and sor-3(bp185) homozygotes show the same PVD mutant phenotype as otDf575 and otDf642 mutant animals (68% penetrant; n=22). sor-3 codes for a protein containing an MBT repeat domain that displays methylated histone binding activity and exists in the PcG proteins SCM and Sfmbt in other organisms (Yang et al., 2007). Previous work showed that loss of sor-3 leads to expression of ectopic dopaminergic and serotonergic male ray neuron fates (Yang et al., 2007). Furthermore, in sor-3 mutants, the Hox genes egl-5 and lin-39 are ectopically expressed outside their usual domain (Yang et al., 2007). Ectopic expression of the mab-5 gene has previously been shown to eliminate the production of the entire postdeirid lineage (that produces the PVD neuron)(Salser and Kenyon, 1996). We hypothesize that sor-3 mutants affect PVD neuron differentiation via ectopic expression of endogenous mab-3.
Reagents
Reagents
*OH9716 otDf575; otIs138; vsIs33
OH9900 otIs138; vsIs33
*OH10091 otDf642; otIs138; vsIs33
*Strains will be available at the CGC.
Acknowledgments
We thank Benjamin Kaplan for technical help.
References
Funding
This work was funded by the Howard Hughes Medical Institute. G.M. was supported by a F31 predoctoral fellowship (1F31NS074841-01).
Reviewed By
Harold SmithHistory
Received: October 17, 2019Accepted: October 23, 2019
Published: October 25, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Minevich, G; Bernstein, A; Mei, K; Poole, RJ; Hobert, O (2019). Nibbling 405 kb off the X: Viable deletion alleles eliminating 50 protein coding genes, including a chromatin factor involved in neuronal development. microPublication Biology. 10.17912/micropub.biology.000187. Corrigendum in: microPublication Biology. 10.17912/micropub.biology.000206.Download: RIS BibTeX