Present address: ARC Centre of Excellence in Plant Energy Biology, The School of Agriculture, Food and Wine, University of Adelaide, Waite Campus, Adelaide, South Australia 5005, Australia
Description
In eukaryotes, RNA and chromatin-based pathways control transposable elements (TE) to minimize the deleterious consequences of genetic invasion, transposition, mutation and chromosome instability (Matzke and Mosher, 2014). In higher plants, the multi-subunit nuclear RNA polymerase IV (Pol IV) specializes in transcribing the 24 nucleotide class of small RNAs that target TE’s for DNA cytosine methylation and silencing in the RNA-directed DNA methylation (RdDM) pathway (Herr et al., 2005; Onodera et al., 2005). In Arabidopsis, the Pol IV has two alternative subunits encoded by the NRPD1a and NRPD1b and a common subunit encoded by NRPD2A (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005). The null mutant for NRPD1a is defective in the RdDM pathway and also displays a late flowering phenotype under short day conditions (Pontier et al., 2005; Eamens et al., 2008).
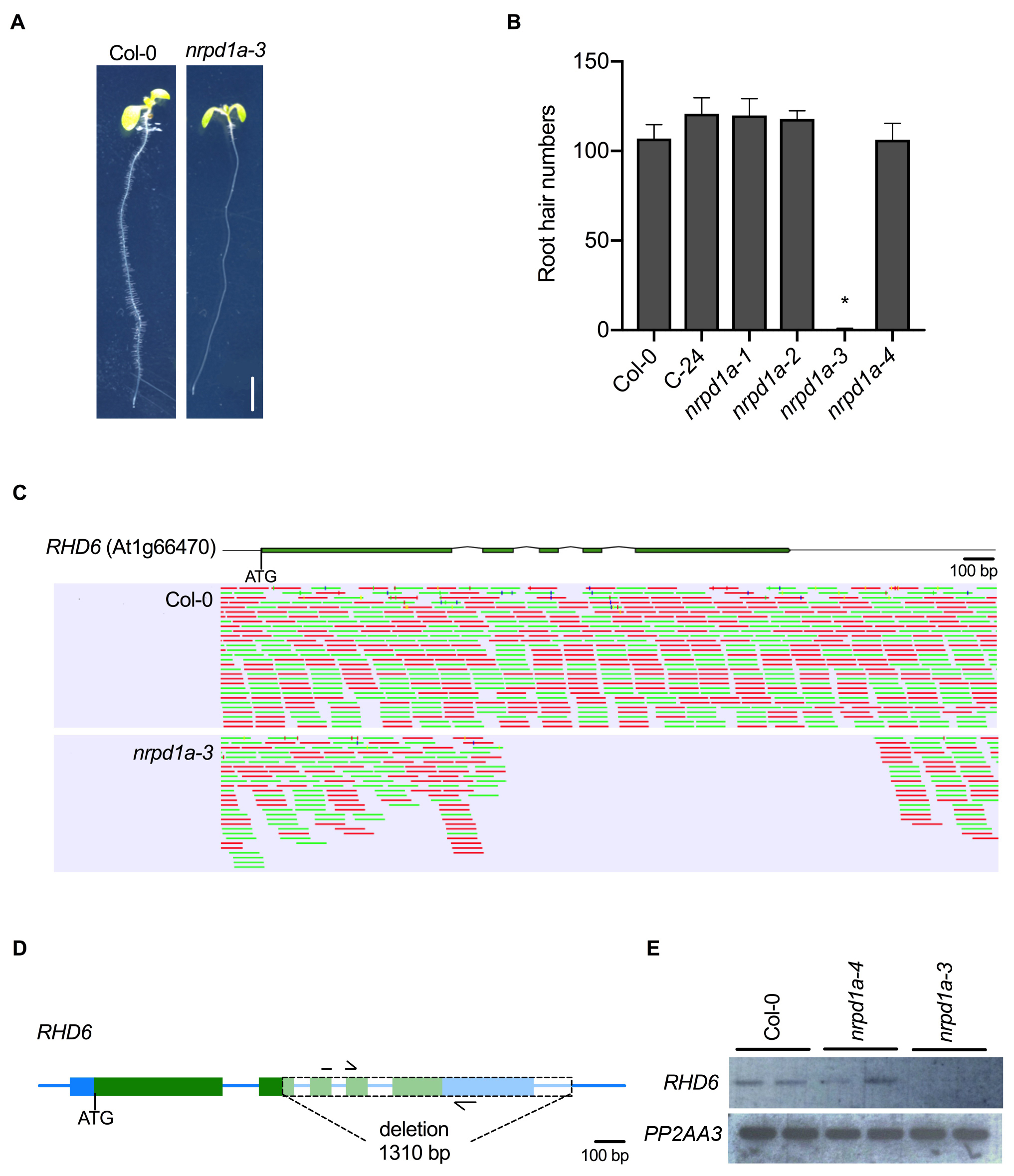
Interestingly, we observed a root hair defective phenotype in the nrpd1a-3 mutant allele which has not been previously reported (Fig. 1A). When grown on vertically oriented agar medium, nrpd1a-3 seedlings lacked root hairs or defective root hair elongation when compared to the Col-0 wild type control which showed normal root hair distribution and length in the root maturation zone (Fig. 1A). Further investigation of other NRPD1A mutant alleles, nrpd1a-1, nrpd1a-2, in the C-24 accession, and nrpd1a-4 in the Col-0 accession revealed that these alleles had a similar root hair number to the wild type controls (Fig. 1B). To investigate if the root hair defective phenotype in nrpd1a-3 was a spurious event in our laboratory’s seed stock or an earlier event, we re-ordered the same mutant from the ABRC stock Centre and tested the root hair phenotype. We observed the same root hair defective phenotype in both nrpd1a-3 seed stocks. We next crossed the nrpd1a-3 mutant (Col-0) to wild type accession C-24, self-fertilized the F1to create a F2mapping population and mapped polymorphic DNA markers across the 5 chromosomes revealed the gene for the root hair defective phenotype was located between DNA markers ciw3 and F26B6 on chromosome 1 (Berendzen et al., 2005). Next we whole-genome sequenced DNA from both Col-0 and nrpd1a-3 using Illumina short read technology, and after GATK (McKenna et al., 2010) and DELLY (Rausch et al., 2012) analysis of the annotated gene models in the genetic window, we identified only one nucleotide mutation, a 1,310 bp deletion in Root Hair Defective 6 (RHD6), in nrpd1a-3 (Fig. 1C). RHD6 is a bHLH transcription factor that positively regulates root hair initiation (Masucci and Schiefelbein, 1994) and loss of function mutations cause a root hair defective phenotype. Semi-quantitative RT-PCR showed RHD6 mRNA was undetectable in the nrpd1a-3 roots when compared to Col-0 wild-type and nrpd1a-4 suggesting that the loss of RHD6 was the likely candidate for the root hair defective phenotype observed in the nrpd1a-3 mutant (Fig. 1D). We confirmed the 1,310 bp deletion that deleted part of exon 2, all of exons 3-5 and part of the 3’ UTR by PCR and Sanger sequencing. Together the genetic mapping and the undetectable RHD6 mRNA transcript strongly suggests that the root hair defective phenotype only observed in the nrpd1a-3 mutant background is caused by the deletion in RHD6. In the Arabidopsis research community, sometimes phenotypes caused by an unlinked mutation to a gene of interest have been incorrectly associated in the research field for many years (Enders et al., 2015; Habets and Offringa, 2015), and so our discovery of a deletion in RHD6 in nrpd1a-3 will allow the community to not incorrectly associate the defective root hair phenotype with POLIV function.
Reagents
For soil grown A. thaliana plants, seeds were germinated in soil and seedlings grown under halogen lights at 21°C under 16-hour light (130 µmoles/m2/sec) and 8-hour dark conditions as previously described (David et al., 2017). For vertical plate experiments, seeds were chlorine gas sterilized as previously described (Burgess et al., 2015) stratified at 4°C in the darkness and then transferred to grow under halogen lights of 16-hour light, 8-hour night conditions (130 µmoles/m2/sec). Seedlings were grown in a vertical orientation for 7 days on half-strength Murashige and Skoog (MS) medium containing 1% sucrose. Col-0 or C-24 accessions were used as wild-type controls. The NRPD1A two SALK T-DNA insertion lines in the Col-0 accessions for NRPD1A were nrpd1a-3 (SALK_128428) and nrpd1a-4 (SALK_083051). Mutants nrpd1a-1 and nrpd1a-2 are in the C-24 accession (Herr et al., 2005). ImageJ was used to measure the number of root hairs longer than 2 mm from an image. Statistical analyses of the data were made using Student’s t-test.
RT-PCR and PCR
Total RNA was isolated from seedling roots using the Tri Reagent (Sigma Aldrich) as described by Wang et al., 2017. Briefly, cDNA for semi-quantitative RT-PCR was synthesized using Superscript III kits as per the manufacturer’s recommendation (Invitrogen) from 2 mg of total RNA that was oligo(dT) primed. Genomic DNA for PCR or sequencing was isolated using the Dellaporta procedure (Dellaporta et al., 1983) with small modifications as previously described from two-week-old seedlings grown on agar medium. RT-PCR and PCR reagents and thermal cycling conditions were previously described in David et al., 2017.
| Gene name | Locus identifier | Application | Forward primer sequence | Reverse primer sequence |
| RHD6 | AT1G14920 | RT-PCR | CCAATGGCACCAAGGTTGATTT | TTTCCCCCGATATTATTACAACGTA |
| PP2AA3 | AT1G13320 | RT-PCR control | GGGCAATGCAGCATATAGTTC | TGGGTCTTCACTTAGCTCCAC |
| RHD6 | AT1G66470 | detection of deletion | AGGGCAACAACATGAGCTACGGC | TAAGAACACGTATCCCTAAT |
Whole-genome sequencing and Bioinformatic analysis
Illumina sequencing libraries were prepared using NEBNext Ultra DNA library Prep kit as per the manufacturer’s recommendation and sequenced on the Illumina Hi-Seq system. Illumina sequence reads were trimmed by using TrimGalore! (Krueger, 2015), sequence quality assessed by using FastQC (Andrews, 2010), detection of nucleotide variation was performed by using DELLY(Rausch et al., 2012)and GATK (McKenna et al., 2010). All bioinformatic analysis were performed using default parameters. Sequence data from this article can be found in the SRA accession ID: PRJNA590836.
Gene sequence data from this article can be found in The Arabidopsis Information Resource (TAIR) under the following accession numbers: NRPD1A At1g63020; RHD6 At1g66470 and PP2AA3 At1g13320.
The nrpd1a-3 rhd6-4 strain has been submitted to ABRC (Stock ID CS72356).
Acknowledgments
We thank the ACRF Cancer Genomics (Adelaide, Australia) staff for assistance with the genome sequencing.
References
Funding
This research was supported by Australian Research Council grant DP190101303.
Reviewed By
AnonymousHistory
Received: November 20, 2019Accepted: December 5, 2019
Published: December 12, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
David, R; Kortschak, RD; Searle, I (2019). The root hair defective phenotype of Arabidopsis thaliana Pol IV subunit mutant nrpd1a-3 is associated with a deletion in RHD6. microPublication Biology. 10.17912/micropub.biology.000196.Download: RIS BibTeX