Description
Human TMEM258 is involved in stress pathways and has been implicated in inflammatory intestinal disorders (Graham et al., 2016). The Drosophila ortholog Kuduk consists of a transmembrane and an intramembrane domain and localizes to the outer nuclear membrane, where it regulates myonuclear positioning and morphology (Ding et al., 2017). Kuduk is the first characterized cytoplasmic regulator of the LINC (linker of nucleoskeleton and cytoskeleton) complex. LINC complexes are conserved throughout eukaryotes and consist of SUN (Sad-1 and UNC-84) proteins, which localize to the inner nuclear membrane and bind to lamin, and KASH proteins (Klarsicht, ANC-1, Syne homology) that span the outer nuclear membrane and interact with cytoskeletal elements. KASH and SUN bind in the perinuclear space to bridge the nucleus and cytoskeleton and transfer force across the nuclear envelope (Starr, 2009). Drosophila Kuduk has been shown to help anchor the nucleus in place via the LINC complex, but the molecular mechanisms it uses to carry out nuclear migration remain unknown.
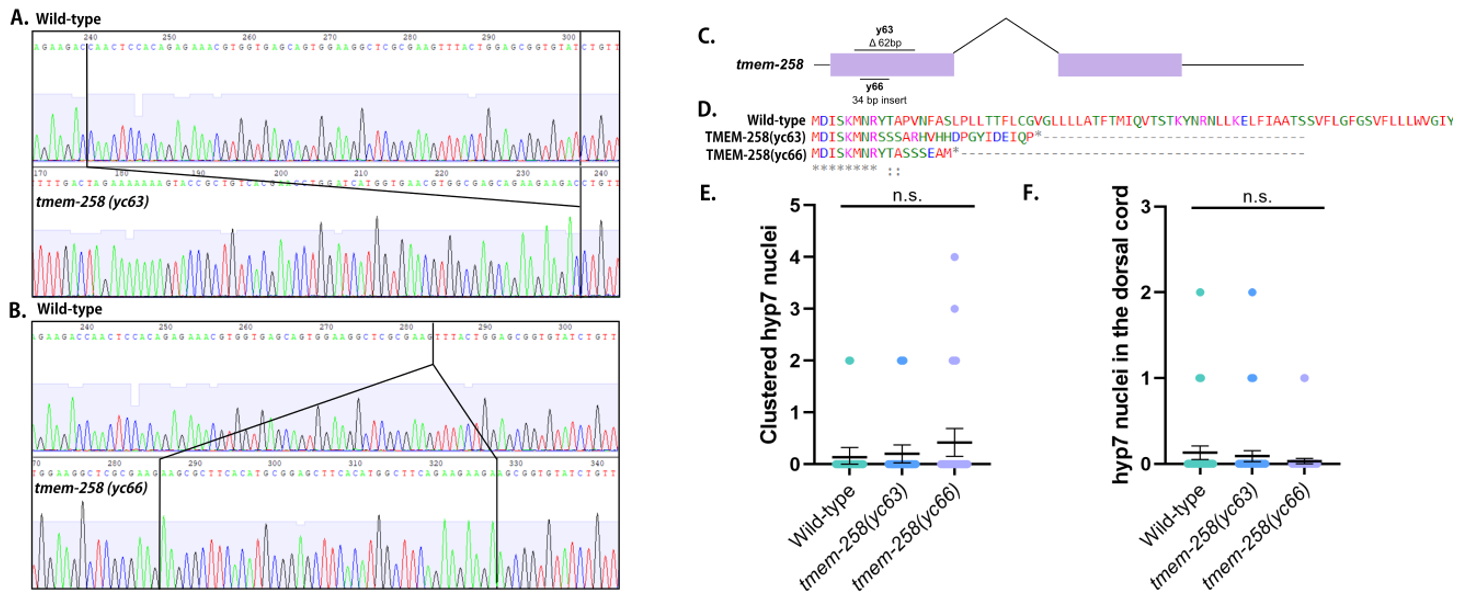
We hypothesized that tmem-258 (formally Y57E12AM.1) is required for nuclear positioning and migration in C. elegans. We used CRISPR-Cas9 to introduce non-specific mutations in tmem-258 and generated two frameshift mutants, yc63 and yc66, which created early stop codons (Fig. 1A-D). We then assayed nuclear migration in two different tissues. First, nuclei migrate across the dorsal midline in hypodermal (hyp7) precursors during normal C. elegans embryogenesis (Fridolfsson et al., 2018). If hyp7 nuclear migration fails, ectopic nuclei can be seen in the dorsal cord. For example, in an unc-84(n369) null line, an average of 15.0 nuclei were abnormally in the dorsal cord (Malone et al 1999). Neither tmem-258(yc63) nor tmem-258(yc66) animals exhibited mislocalized nuclei, suggesting hyp7 nuclear migration was successful (Fig. 1F). Second, we quantified P-cell nuclear migration in L1 larvae. P-cell nuclei migrate from the lateral to the ventral side of the worm where they divide and differentiate to form the vulva and GABA neurons. At 25˚C, unc-84(n369) mutants typically are missing 6.5±0.60 GABA neurons (Bone et al., 2016). To determine whether tmem-258 mutants enhance the P-cell nuclear migration defect of unc-84, we crossed tmem-258(yc63) worms to unc-84(n369) and quantified P-cell nuclear migration failure at 15°C by counting GABA neurons with UNC-47::GFP. The tmem-258(yc63); unc-84(n369) double mutant were missing an average of 0.70±0.3 (mean ± 95% CI, n=25) GABA neurons, equal to the 0.60±0.4 GABA neurons of the unc-84(n369) single mutant at 15˚C. Finally, we tested whether tmem-258 is required for nuclear anchorage in C. elegans by counting the number of syncytial hyp7 nuclei clustering together in adults. The tmem-258 mutants had no significant nuclear anchorage defects (Fig. 1E). We therefore conclude that tmem-258 is not required for nuclear positioning in C. elegans hyp7 cells.
Reagents
The tmem-258 mutations were generated using the guide sequence 5’-TCGCGAAGTTTACTGGAG-3’ injected into UD398: him-8 (e1489) ycIs10 [pBS sk+; pSL589] animals containing a hyp7 GFP nuclear marker. Injections were done with dpy-10 co-CRISPR according to previous methods (Paix et al., 2015). tmem-258(yc63) corresponds to UD600 and tmem-258(yc66) is UD609. tmem-258(yc63) was crossed to UD87: unc-84(n369); oxIs12[unc-47::gfp, dpy-20(+)]; ycEx60[odr-1::rfp] to create UD623: tmem-258(yc63), him-8(e1489) ycIs10; unc-84 (n369) oxIs12[unc-47::gfp] ycEx60[odr-1::rfp unc-84(+)] which was used to assay P-cell nuclear migration.
Acknowledgments
Some strains were supplied by the CGC, which is funded by NIH Office of Research Infrastructure Programs P40 OD010440. We thank the members of the Starr lab for helpful discussions throughout the project.
References
Funding
This study was funded by NIH grant 1R01GM073874.
Reviewed By
David SherwoodHistory
Received: December 12, 2019Accepted: December 31, 2019
Published: January 2, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Gregory, EF; Starr, DA (2020). tmem-258 is dispensable for both nuclear anchorage and migration in C. elegans. microPublication Biology. 10.17912/micropub.biology.000208.Download: RIS BibTeX