Description
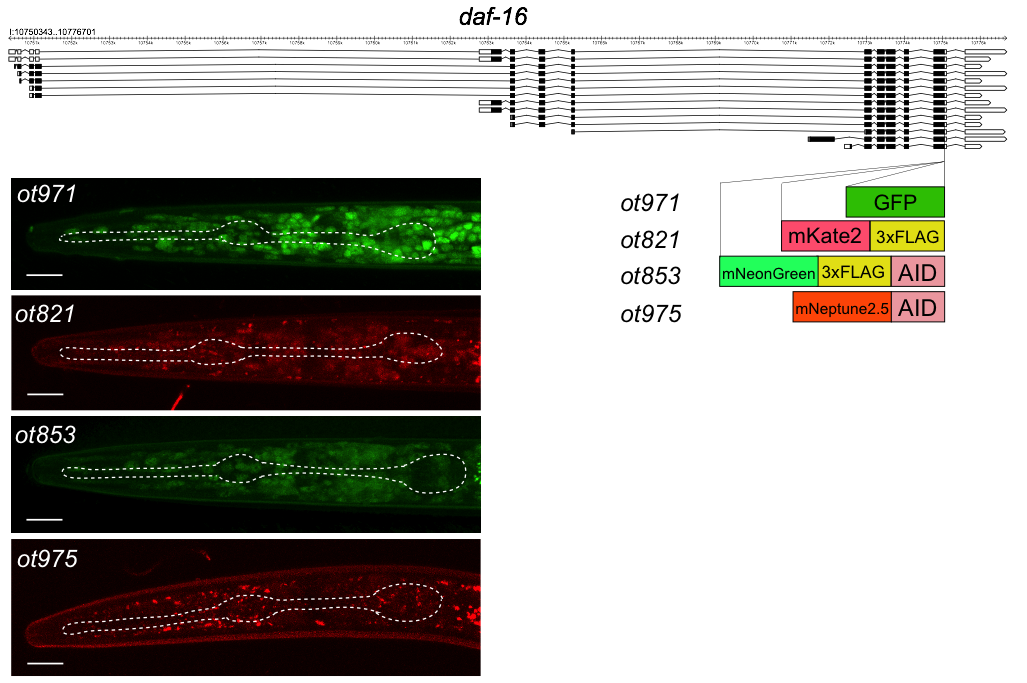
daf-16 encodes a broadly expressed transcription factor that functions in a wide variety of developmental and physiological processes (Lin et al., 1997; Ogg et al., 1997; Tissenbaum, 2018), including in the nervous system (Kim and Webb, 2017). DAF-16 protein shows highly dynamic cytoplasmic to nuclear translocation, which, in the past, has been visualized using multicopy constructs, which can produce potential overexpression artifacts (such as those described in (Henderson and Johnson, 2001)). To avoid such overexpression effects, the generation of fluorescently tagged daf-16 allele would be useful. Similarly, the generation of conditional alleles of daf-16 would be useful to resolve a number of open questions in regard to the focus of daf-16 action. To address both issues, we recently generated an mNeonGreen tagged daf-16 allele that also contained an auxin-inducible degron (Bhattacharya et al., 2019; Zhang et al., 2015). This allele, daf-16(ot853[daf-16::mNG::AID]), enabled us to provide a proof of concept for neuron-type specific daf-16 depletion (Bhattacharya et al., 2019). One issue with this allele has been that due to the emission spectrum of its fluorescent tag (mNeonGreen), it cannot be used in conjunction with gfp-based phenotypic read-outs.
We have now genome-engineered three additional fluorophore tagged daf-16 alleles, some with or without additional tags (FLAG tag or AID degron) (Fig.1). Like the previously published ot853 allele, we CRISPR/Cas9-engineered the ot821 allele (mKate2 tag) with the SEC method (Dickinson et al., 2015) and the ot971 (gfp tag) and ot975 (mNeptune2.5::AID) alleles with the so-called Mello method (Dokshin et al., 2018), using pDD282 (Dickinson et al. 2015) and pEY56 (kindly provided by Eviatar Yemini, based on Addgene plasmid #51310 pcDNA3-mNeptune2.5) as templates. All four alleles show similar expression and subcellular localization patterns. In Figure 1 we show the nuclear localized DAF-16 protein in dauers, generated by a standard crowding/starvation protocol (as described in Bhattacharya et al., 2019).
We note that this tagging approach also provides a rare opportunity to assess the intensities of fluorophores side-by-side: We find that GFP expression is brighter than that of mNeonGreen, while mKate2 is clearly weaker and mNeptune2.5 even weaker.
All strains are available at the CGC.
Reagents
OH16024: daf-16(ot971[daf-16::gfp])
OH13908:daf-16(ot821[daf-16::mKate2::3xFlag])
OH14125:daf-16(ot853[daf-16::mNG::AID])
OH16029: daf-16(ot975[daf-16::mNeptune2.5::AID])
Acknowledgments
We thank Chi Chen for expert assistance in nematode injections and Ev Yemini for providing the mNeptune2.5 template plasmid.
References
Funding
This work was funded by a grant from the NINDS (1 R21 NS115442-01)
Reviewed By
Patrick HuHistory
Received: January 1, 2020Accepted: January 6, 2020
Published: January 7, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Aghayeva, U; Bhattacharya, A; Hobert, O (2020). A panel of fluorophore-tagged daf-16 alleles. microPublication Biology. 10.17912/micropub.biology.000210.Download: RIS BibTeX