Department of MCD Biology, University of California, Santa Cruz, Santa Cruz, CA
Description
The auxin inducible degron (AID) system was developed to enable conditional depletion of proteins of interest. In this system, a plant-derived F-box protein, TIR1, is expressed and complexes with endogenous Skp1 and Cullin proteins to form an E3 ubiquitin ligase complex. Proteins of interest can be tagged with a plant-derived degron, which is recognized by the TIR1-ubiquitin ligase complex in the presence of the plant hormone auxin (indole-3-acetic acid) (Nishimura et al. 2009). This system was recently adapted for Caenorhabditis elegans, wherein TIR1 constitutive expression enables rapid depletion of degron-tagged proteins of interest when animals are transferred to NGM plates supplemented with auxin. In C. elegans, the AID system has allowed for fine-tuned spatiotemporal control of protein expression and has been used to deplete many essential proteins, which are difficult to study using classical genetic approaches (Zhang et al. 2015). Here, we highlight a limitation of this system by demonstrating that TIR1 expression can cause depletion of degron-tagged proteins without auxin addition. These results corroborate observations from human cell lines, where TIR1 has been shown to act independently of auxin to chronically deplete degron-tagged proteins (Sathyan et al. 2019).
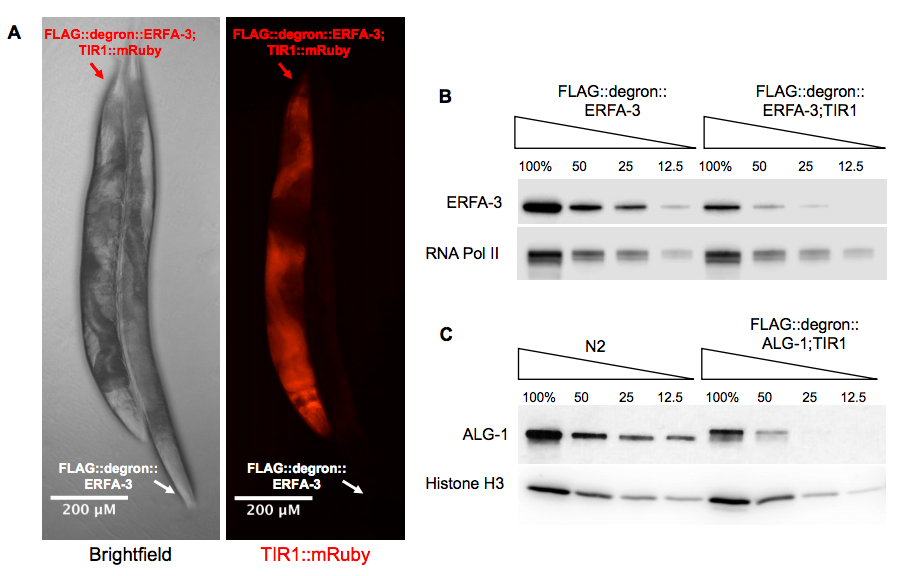
In an attempt to study the effects of depletion of the essential protein ERFA-3, we used CRISPR-Cas9 to insert a degron sequence at its N-terminus. The resulting 3xflag::degron::erfa-3 strain (WJA2126), was backcrossed three times to N2 and then outcrossed to CA1200, a publicly available pan-somatic TIR1::mRuby strain (ieSi57 [eft-3p::TIR1::mRuby::unc-54(3’UTR) + Cbr-unc-119(+)] II), to make a new strain, PQ631. Expression of TIR1 in the 3xflag::degron::erfa-3 strain led to gross morphological defects that are not seen in the 3xflag::degron::erfa-3 or TIR1-expressing strains (Zhang et al. 2015) (Figure 1A). The 3xflag::degron::erfa-3; ieSi57 [Peft-3::TIR1::mRuby::unc-54 3’UTR, cb-unc-119(+)] II strain exhibited bagging and dumpy phenotypes as well as fertility defects that manifested as an observed slower growth rate of the overall population in comparison to its parental strains. Individuals of this strain tended to lay fewer viable eggs than control strains. It is yet unclear whether this is due to defects in meiosis, embryogenesis, or in any of the numerous steps in-between. To understand the cause of these phenotypic abnormalities, we performed a quantitative Western blot against ERFA-3 using an anti-FLAG antibody (Figure 1B). Here, it is evident that expression of TIR1 alone, despite no addition of auxin to the media, leads to depletion of 3xFLAG::degron::ERFA-3 to approximately 30% of the levels observed in the absence of TIR1.
We observed a similar phenomenon with the alg-1 locus. We analyzed QK155 (4xflag::degron::alg-1; ieSi57 [eft-3p::TIR1::mRuby::unc-54(3’UTR) + Cbr-unc-119(+)] II). This strain expresses degron-tagged ALG-1 and pan-somatic TIR1. We performed a quantitative Western blot to compare ALG-1 expression in our 4xFLAG::degron::ALG-1; TIR1 strain to N2 using an antibody against endogenous ALG-1. As with ERFA-3, degron-tagged ALG-1 was chronically depleted when TIR1 was expressed pan-somatically. The ALG-1 protein level in the 4xFLAG::degron::ALG-1;TIR1 strain was approximately 25% that of endogenous protein expression (Figure 1C).
Taken together, these results indicate that TIR1 can act without the addition of auxin to the media, ultimately resulting in chronic depletion of degron-tagged proteins. There are at least two, non-mutually exclusive possibilities to explain our observations: (1) Auxin may exist in one or more components of the C. elegans media used. Seaweed is a common source of agar and does contain some auxin, though it is unclear if the concentration would be sufficient to activate TIR1. (2) There may be auxin-independent recognition of the degron by TIR1. Regardless of the mechanism of our observation, the effects were observed in at least two separate labs over the span of several months, indicating it is a potentially widespread caveat of the approach.
The phenotypes we observed for the 3xflag::degron::erfa-3 strain in the presence of TIR1 are consistent with published phenotypes associated with RNAi knockdown of erfa-3, which include sterility, slow development, and a high rate of embryonic lethality (Kamath et al. 2003). Yet, when the AID system was adapted for C. elegans, various phenotypic assays were performed to demonstrate that TIR1 expression, with multiple different degron-tagged essential proteins, resulted in no observable abnormalities (Zhang et al. 2015). This suggests that not all degron-tagged proteins may be subject to auxin-independent degradation by TIR1 or that chronically lower levels of some essential proteins can be tolerated. In conclusion, our results highlight the need to carefully analyze expression levels of degron-tagged proteins of interest as they may be subject to TIR1-mediated degradation prior to auxin exposure.
Methods
Request a detailed protocolImaging: WJA2126 and PQ631 animals were cultured at 15°C and grown until adulthood before being anesthetized with 1 mg/ ml of Levamisole and imaged at 10X magnification. Imaging was performed with Zeiss Axio Imager.A1.
Western blotting: Western blotting was performed as previously described (Zisoulis et al. 2010; Van Wynsberghe et al. 2011), using mouse monoclonal antibodies against FLAG (Sigma) and RNA polymerase II (Covance) or rabbit polyclonal antibodies against Histone H3 (Abcam) and ALG-1 (ThermoFisher Scientific).
Reagents
Nematode culture: C. elegans strains were cultured under standard conditions and synchronized by hypochlorite treatment (Wood 1988).
Strain construction: erfa-3 was tagged with 3xflag::degron by CRISPR/Cas9 using the co-conversion strategy as previously described (Arribere et al. 2014) using overlapping ultramers (IDT) encoding the tag as outlined previously (Paix et al. 2016). Integration at the erfa-3 locus was confirmed by sequencing. The sequence of the insertion with flanking homologies is aagcgatttcagagctctcggcatcgacgcaaaATGTCCgattataaagatcatgacggagattataaagaccatgatattgatt
ataaagatgacgatgataagATGCCTAAAGATCCAGCCAAACCTCCGGCCAAGGCACAAGTTGTGGGATGGCCACCGGTGAGATC
ATACCGGAAGAACGTGATGGTTTCCTGCCAAAAATCAAGCGGTGGCCCGGAGGCGGCGGCGTTCGTGAAGTCAGGCTGGAACGTG
AACGCCTCGTCGTTTGTGCCAAA.
Strains:
WJA2126: erfa-3(srf2126 | 3xflag::degron::erfa-3) V, this study
CA1200: ieSi57 [eft-3p::TIR1::mRuby::unc-54(3’UTR) + Cbr-unc-119(+)] II, CGC (The eft-3 gene is also referred to as eef-1A.1.)
PQ631: erfa-3(srf2126) V; ieSi57 II, this study
QK155: alg-1(xk20 | 4xflag::degron::alg-1) X; ieSi57 II, gift from the John Kim Lab
Acknowledgments
We thank Shreya Singireddy, Himani Galagali and John Kim for sharing the QK155 strain.
References
Funding
Support for this work was from the UCSD Cellular and Molecular Genetics Training Program through an institutional grant from the National Institute of General Medicine (T32 GM007240) to E.C.S. and A.L.N., a National Science Foundation Graduate Research Fellowship (DGE-1650112) to A.L.N., a grant from the National Institute of General Medicine (R35 GM127012) to A.E.P., a grant from the National Institute of General Medicine (R01 GM131012) to J.A.A., start-up funds from UCSC to J.A.A., a grant from the Searles Scholar Foundation to J.A.A., and the UCSC Initiative for Maximizing Student Development through an institutional grant from the National Institute of General Medical Sciences (R25 GM058903) to M.S.M.
Reviewed By
Scott KennedyHistory
Received: December 20, 2019Accepted: January 7, 2020
Published: January 28, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Schiksnis, EC; Nicholson, AL; Modena, MS; Pule, MN; Arribere, JA; Pasquinelli, AE (2020). Auxin-independent depletion of degron-tagged proteins by TIR1. microPublication Biology. 10.17912/micropub.biology.000213.Download: RIS BibTeX