Department of Biotechnology, Toyama Prefectural University, 5180 Kurokawa, Imizu, Toyama 939-0398, Japan
Department of Mechanical Engineering, School of Engineering, Tokai University, 4-1-1 Kitakaname, Hiratsuka, Kanagawa 259-1292, Japan
Description
OP50 is an Escherichia coli strain conventionally used as a bacterial food in the laboratory maintenance of Caenorhabditis elegans on agar plates. It has also been used to feed C. elegans in longitudinal cultures within microfluidic devices (MFDs) (Hulme et al., 2010; Li et al., 2015), where it has been subject to killing by ultraviolet irradiation or pasteurization performed to suppress clogging due to biofilm formation and aggregation (Li et al., 2015; Zhuo et al., 2017). However, the killed bacterial food can change C. elegans aging dynamics, likely due to influences on C. elegans physiology (Saul et al., 2009; Gruber et al.;, 2007; Garigan et al., 2002). Further development of longitudinal culturing systems for C. elegans in MFDs requires elucidation of the mechanisms that underlie food bacteria clogging and delineation of culture conditions in which living bacterial food can be incorporated without clogging. Bacteria switch from planktonic growth to aggregated growth under conditions of environmental stress, in the presence of toxins (e.g. antibiotics), and when there is a lack of nutrients (Trunk et al., 2018). Biofilms, such as dental plaque, are bacterial communities that are organized in a film-like form in which they are embedded in a self-produced polymeric matrix on biotic or abiotic surfaces; pellicles are floating biofilms that form at liquid-air interfaces. Meanwhile, autoaggregations are aggregated communities of bacteria suspended in solution, such as bacterial flocs formed in activated sludge. Biofilms and autoaggregations are formed by both shared and independent genetic and physico-chemical mechanisms (Trunk et al., 2018; Berne et al., 2018; Berne et al., 2015). In this study, we examined OP50 biofilm formation.
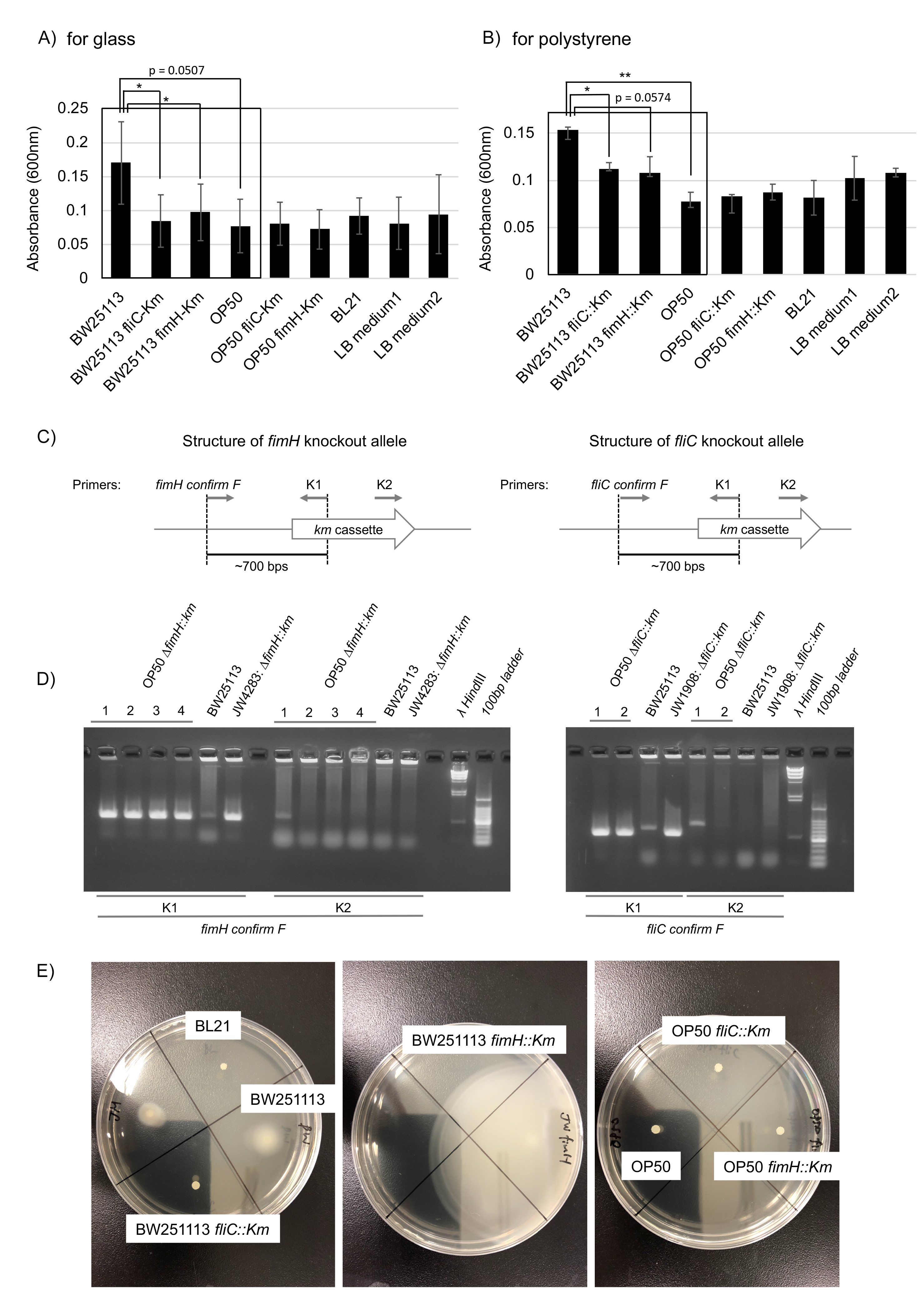
Biofilm formation is mediated by flagellin proteins (e.g. FliC), which form flagella, and the adhesion protein FimH, which is located at the tips of type I pili (Berne et al., 2018, Jones et al., 1995; Pratt and Kolter, 1998; Friedlander et al., 2013). We compared the biofilm formation ability of OP50 with that of the biofilm-forming (Wood et al., 2006) wild-type BW251113 E. coli strain as well as that of two BW251113-derived knockouts produced with a kanamycin (Km) cassette characterized as biofilm formation defective mutants: JW4283: BW25113 fimH::Km (a fimH knockout) and JW1908: BW25113 fliC::Km (a fliC knockout) (Baba et al., 2006). Compared to the original BW251113 strain, BW251113 fliC::Km had a significantly reduced ability to form biofilm on glass and polystyrene (Fig. 1A and 1B, p < 0.05) and BW25113 fimH::Km had a significantly reduced ability to form biofilm on glass (Fig. 1A, p < 0.05; biofilm formation on polystyrene showed a near-significant reduction trend Fig. 1B, p = 0.0574). Compared with the original BW251113 strain, we found that OP50 had a significantly reduced biofilm formation ability on polystyrene (Fig. 1B, p < 0.05; biofilm formation on glass showed a near-significant reduction trend, Fig. 1A, p = 0.0507). The biofilm formation ability of OP50 was as low as that seen with the BW251113 biofilm formation defective mutants, and similar to that of OP50 fliC::Km and OP50 fimH::Km mutants (Fig. 1A and 1B), which were constructed by transferring fliC::Km and fimH::Km alleles to OP50 by P1 transduction (Fig. 1C and 1D). Therefore, we conclude that the original OP50 strain is itself a biofilm formation defective mutant.
Bacterial motility is required for biofilm formation. Despite of the presence of flagella, mutants of a motor protein that enables flagellar rotation have been shown to have a reduced biofilm formation ability (Pratt and Kolter, 1998; Friedlander et al., 2013; Wood et al., 2006). To study the mechanism of impaired biofilm formation ability of OP50, we performed a motility assay that showed that OP50 exhibited much lower motility than BW251113 (Fig. 1E). Motility was similar among the original OP50, OP50 fliC::Km, and OP50 fimH::Km strains (Fig. 1E). Preliminary mapping of a draft genome sequence of OP50 to that of a reference E. coli strain (REL606) identified non-synonymous nucleic acid substitutions in flagellar hook protein E, flagellar P-ring protein precursor I, and flagellar hook-associated protein K (personal communication with Robin C. May at University of Birmingham) (see Excel file in WormBase, ftp://ftp.wormbase.org/pub/wormbase/species/e_coli/op50/annotation/op50_annotated_snps.xls). These results suggest that OP50’s impaired biofilm formation is likely caused by a lack of functional flagella due to these point mutations on flagella genes.
In our experiments, the motility and biofilm formation ability on glass and polystyrene of BL21 strain E. coli was similar to that seen with BW25113 fliC::Km (Fig. 1A, 1B, and 1E). Our preliminary experiments suggest that BL21 is also a biofilm formation defective mutant with impaired motility. Impaired biofilm formation in BL21 and OP50 due to impaired motility is not necessarily caused by maintenance in nutrient-rich laboratory conditions, where bacteria can survive without active chemotaxis by flagella. It has been reported that flagellar regulon is frequently deleted secondarily to experimental genetic manipulation (Hobman et al., 2007). Additionally, E. coli mutants with no functional flagella are selected in mouse intestine under certain conditions (Leatham et al., 2005). These phenomena are thought to be caused by high selective pressure from extracellular nutrient conditions on the flagellum regulon, because flagella are high energy consumers. Given that non-flagellar mutants are frequently selected in response to energy balance conditions, OP50 and BL21 may be ordinary examples of biofilm defective mutants among laboratory E. coli strains.
We found that motility was greatly enhanced in BW25113 fimH::Km, compared to original BW25113 (Fig. 1E). It has been shown that down-regulation of the expression of genes responsible for the formation of pili, including fimH, result in an upregulation of fliC, leading to enhanced motility (Leatham et al., 2005). The high motility of BW25113 fimH::Km is likely caused by anticorrelated control between fimH and fliC. Even in high-motility BW25113 fimH::Km, FimH is required for biofilm formation (Fig. 1A). The lack of motility enhancement in OP50 fimH::Km (Fig. 1E) is likely due to OP50’s non-functional flagella.
Our finding that OP50 is a biofilm formation-deficient mutant suggests that a genetic mechanism of biofilm formation is unlikely to be a direct cause of OP50 clogging in MFDs. Experimentally, autoaggregation has been studied by examining bacterial sedimentation in a culture solution (Trunk et al., 2018). Autoaggregation has been shown to be promoted by Van der Waals forces between adhesion proteins, such as Flu/Ag43 (Lane et al., 2007) and TibA (Hasman et al., 1999), and by depletion forces between bacterial cells that have been defined in colloid physics (Sherlock et al., 2005; Schwarz-Linek et al., 2010; Roux et al., 2005). Thus, it would be prudent to study how MFD clogging is affected by genetic and physical mechanisms of auto-aggregation in efforts to optimize conditions for using living bacteria for food in culture systems in MFDs.
Methods
Request a detailed protocolBiofilm assay
Biofilm was quantified by a crystal violet (CV) (Tokyo Chemical Industry Co., LTD. Japan) quantitation method (Pratt and Kolter, 1998, Roux et al., 2005). A single E. coli strain colony was inoculated in 1 ml LB medium in a polystyrene 24-well plate (Multidish 24 wells Nunclon Delta SI, 142475, Nunc, Denmark, and 24-well Cell Culture Plate, 3524, Corning, USA) or glass tube (IWAKI 10X 100 mm 9832-1310 Asahi Glass Co. LTD., Japan), and cultured at room temperature for 2 d. After cultivation, the plate or glass tube was washed twice with 2 ml phosphate buffered saline and then incubated with 1% CV solution for 1 h. After CV staining, the plate or glass tube was washed with 2 ml phosphate buffered saline, and CV was resolved by 200 µl ethanol:acetone (8:2) solution. CV in ethanol:acetone solution was quantified by optical density at 600 nm (Microplate reader, Epoch2, BioTek, USA).
Isolation of OP50 fliC::Km and OP50 fimH::Km
Km cassettes, fliC::Km and fimH::Km in JW1908: BW25113 fliC::Km or JW4283: BW25113 fimH::Km, were transferred to OP50 by P1 transduction. The presence of fliC::Km and fimH::Km in OP50-derived colonies was checked by polymerase chain reaction (PCR) using a primer set between K1 or K2 primers (Datsenko and Wanner, 2000) and fimH confirming F primer (CACAATCAGCGCACTTCCCGTTACAG) or fliC confirming F primer (AGAAAAGAGTATTTCGGCGACTAAC) with bacteria picked up from a colony of candidates OP50 fimH::Km and OP50 fliC::Km. Bacteria obtained from all colonies of candidate OP50 fimH::Km and OP50 fliC::Km were subjected to PCR with the indicated primers; PCRs amplified DNA fragments of expected size (~700 base pairs) when a primer set containing K1 (but K2) primer was used. We used clone 1 for OP50 fimH::Km and OP50 fliC::Km.
Motility assay
Motility of bacteria was compared by quantitation of colony size after culturing on LB plates with 0.3% agar (Eaves-Piles et al., 2008; Rashid and Kornberg, 2000). One-microliter aliquots of overnight culture solution of LB media were spotted onto soft agar LB plates and incubate for 2 d at room temperature.
Statistical analysis
Statistical testing was performed with paired Student’s t-tests and p-values in multiple comparison were determined after Bonferroni correction. We considered p < 0.05 to be statistically significant.
Reagents
BW25113 fliC::Km and JW4283: BW25113 fimH::Km were obtained from the National Institute of Genetics, Japan.
Acknowledgments
We thank the Caenorhabditis Genetics Center (CGC) for the bacterial strain OP50. The CGC is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the National BioResource Project-E.coli at NIG for E. coli strains.
References
Funding
Y. Arata is supported by a research grant, Challenging Research (Pioneering), Grants-in-Aid for Scientific Research (18H05300).
Reviewed By
AnonymousHistory
Received: January 8, 2020Accepted: January 25, 2020
Published: February 5, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Arata, Y; Oshima, T; Ikeda, Y; Kimura, H; Sako, Y (2020). OP50, a bacterial strain conventionally used as food for laboratory maintenance of C. elegans, is a biofilm formation defective mutant. microPublication Biology. 10.17912/micropub.biology.000216.Download: RIS BibTeX