Biotech Research and Innovation Centre, University of Copenhagen, Ole Maaløes Vej 5, Copenhagen, Denmark
Description
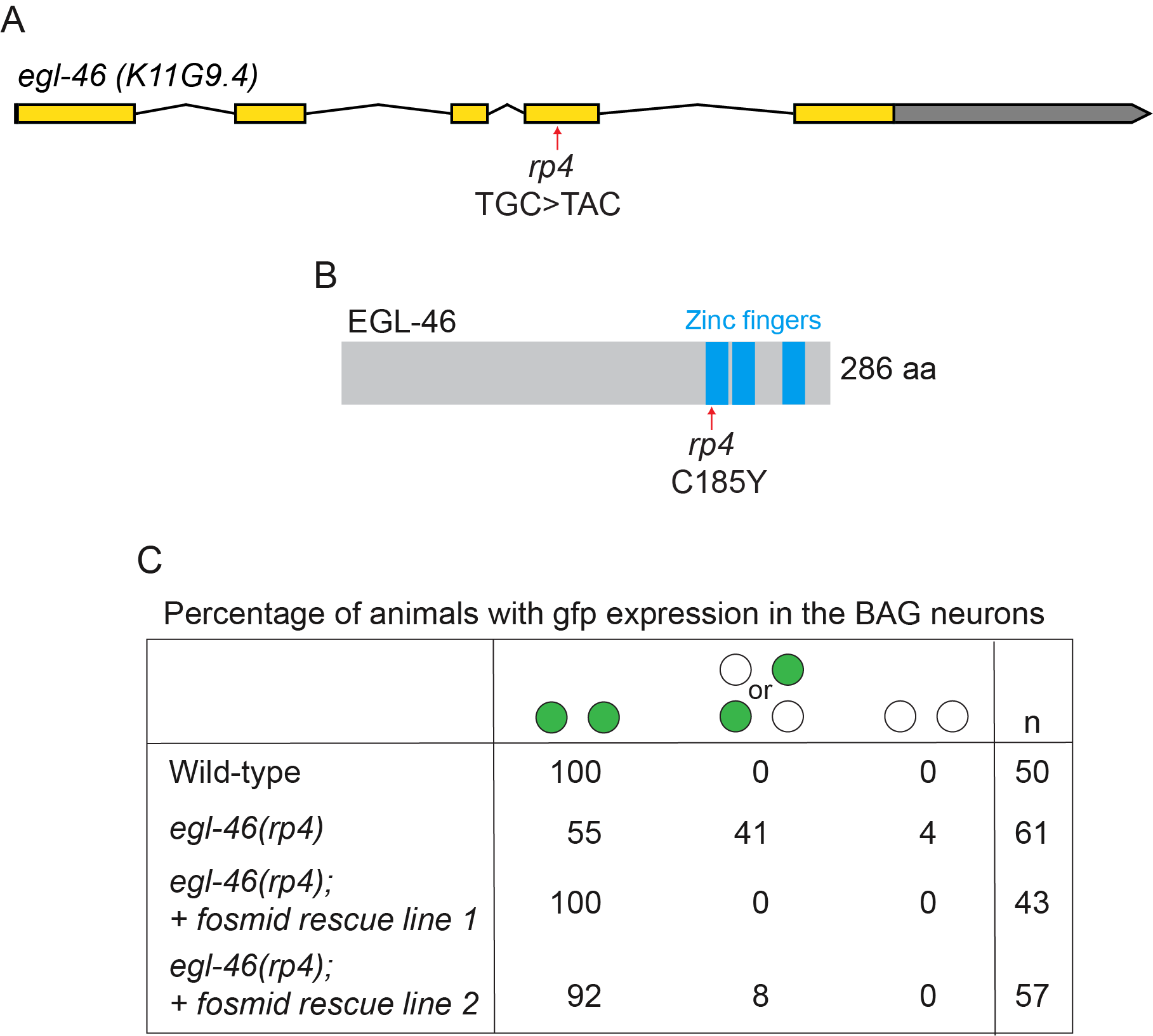
The BAG neurons control multiple aspects of Caenorhabditis elegans behavior, such as sensing environmental gases (oxygen and carbon dioxide), regulation of systemic fat levels and egg laying (Brandt et al. 2012; Guillermin et al. 2011; Juozaityte et al. 2017; Zimmer et al. 2009). To identify factors that control BAG specification, we performed a forward genetic mutagenesis screen using the Pgcy-33::gfp reporter, which is exclusively expressed in the BAG neurons. We isolated a new allele (rp4) that exhibits a loss of Pgcy-33::gfp expression. Using the one-step whole-genome sequencing and SNP mapping strategy (Doitsidou et al. 2010) we mapped the genetic lesion to the egl-46 gene, encoding a zinc finger transcription factor homologous to mammalian INSM1/2, which we had previously shown to be important for BAG specification (Rojo Romanos et al. 2015). The new lesion we identified egl-46(rp4) (TGC>TAC) causes a single amino acid change in a highly conserved cysteine residue (C185Y) that lies in the first zinc finger domain of EGL-46, which would be predicted to affect DNA binding. Analysis of Pgcy-33::gfp expression in the rp4 allele reveals that it exhibits the same phenotype as the previously published rp13 deletion allele, which is an out-of-frame deletion that removes the zinc finger domains (Rojo Romanos et al. 2015). Therefore, rp4 acts as a strong loss-of-function/null allele and may be of use to those researchers interested in elucidating additional functions of EGL-46.
Methods
Request a detailed protocolIn the forward genetic screen, the BAG reporter strain Pgcy-33::gfp; Pdop-3::rfp was mutagenized using ethyl methanesulfonate. Mutants with decreased GFP expression in the BAG neurons were isolated using the automated COPAS biosorter platform. The one-step whole-genome sequencing and SNP mapping strategy (Doitsidou et al. 2010) was used to identify the genetic lesion of the isolated rp4 allele. Phenotypic analysis of Pgcy-33::gfp BAG expression was performed as described previously (Rojo Romanos et al. 2015).
Reagents
RJP22 rpIs3(Pgcy-33::gfp); vsIs33(Pdop-3::rfp)
RJP56 egl-46(rp4); rpIs3(Pgcy-33::gfp); vsIs33(Pdop-3::rfp)
RJP4585 egl-46(rp4); rpIs3(Pgcy-33::gfp); vsIs33(Pdop-3::rfp); rpEx2046 (WRM0636bB06) 1ng/µl + Punc-122::gfp 30ng/µl Line 1
RJP4586 egl-46(rp4); rpIs3(Pgcy-33::gfp); vsIs33(Pdop-3::rfp); rpEx2047 (WRM0636bB06) 1ng/µl + Punc-122::gfp 30ng/µl Line 2
Strains will be available at the CGC.
References
Funding
NHMRC - GNT1105374 and GNT1137645 to R.P.
Reviewed By
David MillerHistory
Received: February 11, 2020Accepted: February 25, 2020
Published: February 25, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Godini, R; Langebeck-Jensen, K; Pocock, R (2020). A single amino acid change in the EGL-46 transcription factor causes defects in BAG neuron specification. microPublication Biology. 10.17912/micropub.biology.000224.Download: RIS BibTeX