Laboratory of Biochemistry and Genetics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD
Presently- Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, TN
Description
The 26S proteasome is one of the major proteolytic machineries in cells and is composed of nearly 33 different subunits (Budenholzer et al., 2017; Chen et al., 2008; Papaevgeniou and Chondrogianni, 2014). The subunits are arranged as one or two 19S regulatory particle(s) capping a cylindrical catalytic 20S core particle (Budenholzer et al., 2017). Each subunit is highly conserved from yeast to mammals, and proper function of the proteasome is crucial for survival of organisms (Papaevgeniou and Chondrogianni, 2014). Spatiotemporal expression of specific subunits and their roles in regulating the proteasome activity is still not clearly understood.
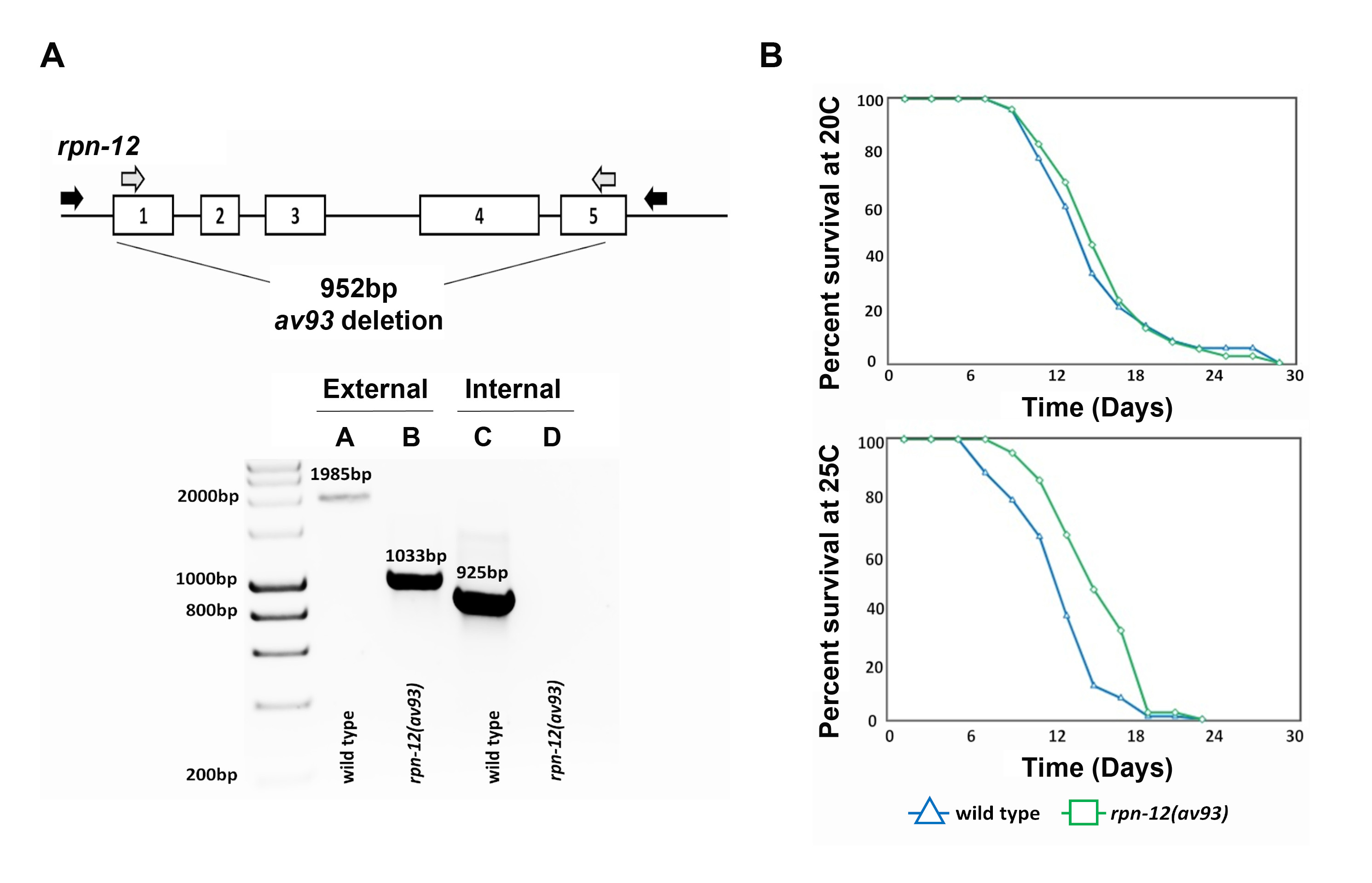
RPN-12 is a subunit of the 19S regulatory particle (RP) of the 26S proteasome in Caenorhabditis elegans (Boehringer et al., 2012; Takahashi et al., 2002). In C. elegans, loss or down regulation of a single proteasome subunit causes embryonic lethality, with the exception of three of the 19S RP subunits: RPN-10, RPN-12, and DSS-1 (Keith et al., 2016; Pispa et al., 2008; Shimada et al., 2006; Takahashi et al., 2002). While the roles of RPN-10 and DSS-1 were previously studied, more detailed investigation into the function of RPN-12 has not been performed. A heterozygous mutant containing a 952bp deletion of the coding region of rpn-12 was generated [rpn-12(av93)] using CRISPR/Cas9 genome editing technology. The heterozygous rpn-12 null mutant was then homozygosed and determined to be homozygous viable with apparent fertility issues. Only 36% of the rpn-12(av93) hermaphrodites produced progeny due to sperm production defects (this phenotype will be published elsewhere). Since efficient proteasome activity is important for the longevity of most organisms, we wanted to investigate whether rpn-12(av93) mutants may have an altered lifespan compared to wild type animals (Saez and Vilchez, 2014; Vilchez et al., 2012). To determine whether the lifespan of rpn-12(av93) mutant animals was affected, lifespan assays were conducted at 20°C and 25°C. The mean lifespan of rpn-12(av93) hermaphrodites (15.94 days) was not significantly different from wild type hermaphrodites (15.54 days) at 20°C. Interestingly, at 25°C the mean lifespan of rpn-12(av93) hermaphrodites (15.59 days) was increased compared to wild type animals (12.80 days). However, the maximum lifespan of both rpn-12(av93) and wild type hermaphrodites was not significantly different at 25°C. The lifespan analysis at 20°C was performed three times (cumulative wild type n=240 and mutant n=192) and at 25°C was performed twice (cumulative wild type n=185 and mutant n=135). Our data suggests that RPN-12 is not essential for viability and lifespan of C. elegans under normal conditions (20ºC), but that absence of RPN-12 can result in a significant increase in mean lifespan under heat stress (25ºC).
Methods
Request a detailed protocolThe rpn-12(av93) AG343 strain was generated via CRISPR/Cas9 genome editing technology following the direct delivery method developed by the Seydoux laboratory (Paix et al., 2017). Two crRNAs (5’- agccagaagatttttatggg – 3’and 5’- actcgaacaaatcgtttaac – 3’) were used to guide the Cas9 to make cuts at specific regions of either ends of the rpn-12 gene. An ssODN (5’-aaacattattggatttaagaaaatgtctgccgcccaaccggtgtctttcaagaactcaagcattgtattt-3’) was delivered as a template for the repair that results in a 952bp deletion that removes most of the first exon and then the entire second to last exons. Screening was performed using the co-conversion dpy-10 method (Arribere et al., 2014). Two independent strains of the rpn-12 deletion were generated [rpn-12(av93) AG343 and rpn-12(ana10) WDC10] through the previously described method, and each strain outcrossed five times before experiments conducted. Both strains showed similar viability and fertility phenotypes, however only rpn-12(av93) was used for the lifespan experiments. Lifespan assays were conducted by placing age synchronized L4 larvae of rpn-12(av93) and wild type animals on MYOB plates seeded with OP50 (10-20 worms per plate). The worms were then passaged to new MYOB plates every two days. The number of dead hermaphrodites was scored every two days until all worms were dead and the percentage of surviving animals calculated. Missing worms were excluded from the statistical analysis.
Reagents
AG343 rpn-12(av93)
WDC10 rpn-12(ana10)
References
Funding
This work was supported, in part, by a grant W911NF-18-1-0465 from the Department of Defense to A.K.A. (V.N., L.M.F. and A.K.A.) and by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (T.J.H. and A.G.).
Reviewed By
AnonymousHistory
Received: July 26, 2019Revision received: March 28, 2020
Accepted: March 30, 2020
Published: April 6, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Fernando, L; Nguyen, V; Hansen, T; Golden, A; Allen, A (2020). Loss of proteasome subunit RPN-12 causes an increased mean lifespan at a higher temperature in C. elegans. microPublication Biology. 10.17912/micropub.biology.000234.Download: RIS BibTeX