Department of Biology, University of Nevada, Reno, Reno, Nevada 89557
Description
Sleep is intertwined with metabolic function in vertebrates (Tsuneki et al. 2016; Herrera et al. 2017; Aalling et al. 2018; Wilms et al. 2018) and invertebrates (Kempf et al. 2019; Ki and Lim 2019; Yurgel et al. 2019; Grubbs et al. 2020), but the molecular underpinnings of this connection are not well understood. We recently reported that the salt inducible kinase (SIK) homolog KIN-29 is required in a subset of sensory neurons for the metabolic regulation of sleep in Caenorhabditis elegans (Grubbs et al. 2020). However, since our genetic manipulations made use of mutations that were present throughout the life of the animal, it is possible that the sleep defect of kin-29 mutants reflects a requirement for KIN-29 activity during development rather than during sleep. Indeed, because kin-29 is expressed throughout development as well as adulthood (Maduzia et al. 2005), and kin-29 mutants show altered expression of targets influential in larval development (Van Der Linden et al. 2008), a role for kin-29 during development remained plausible. Distinguishing a developmental early role from a role during the time of sleep is important for constraining models for how KIN-29 regulates sleep.
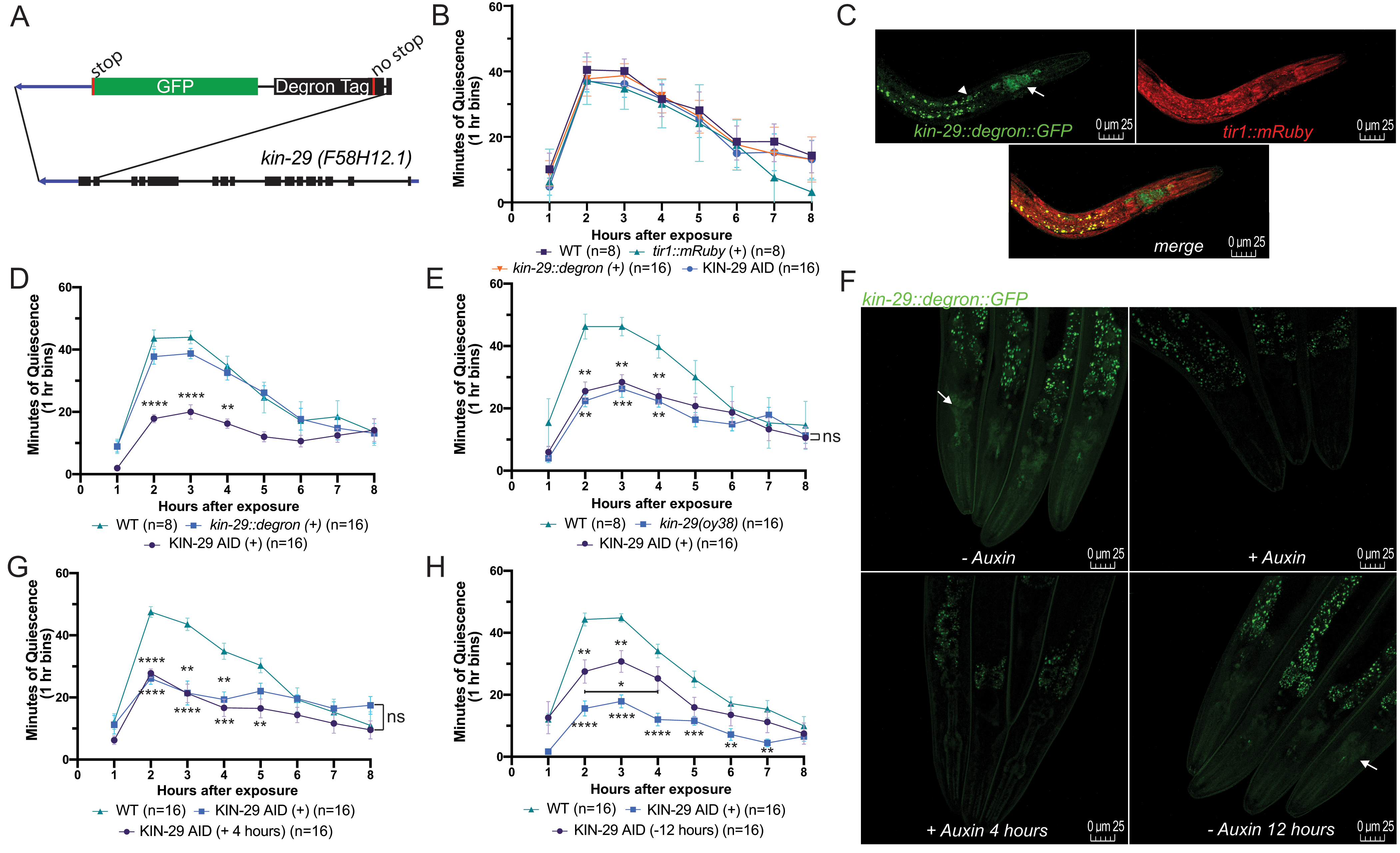
To determine the KIN-29 time window of action, we depleted the KIN-29 protein in a temporally-controlled fashion. We used CRISPR/Cas9 genome editing to introduce a DEGRON fused to GFP just before the kin-29 stop codon (Fig. 1A). After confirming successful editing by Sanger sequencing, we crossed the kin-29::degron::GFP into the previously generated Peft-3::TIR1::mRuby strain (Zhang et al. 2015) to allow for rapid and reversible degradation of KIN-29 when exposed to the phytohormone auxin (Ruegger et al. 1998; Gray et al. 1999). We examined stress-induced sleep (SIS) following exposure to UVC-irradiation (Debardeleben et al. 2017) in the first day of adulthood. We first verified that auxin exposure did not disrupt SIS in the parental strains, and that the progeny containing both transgenes, from here on referred to as KIN-29 Auxin-Inducible Degron (KIN-29 AID) animals, show normal SIS in the absence of auxin (Fig. 1B). We also confirmed the expression of both transgenes in KIN-29 AID animals (Fig. 1C). We then measured SIS in KIN-29 AID transgenic worms cultivated on agar containing auxin (2 mM) from the time of hatching. KIN-29 AID transgenic animals cultivated on auxin throughout development mimicked the kin-29(oy38) null mutant SIS phenotype (Fig. 1D-E). Such animals also showed undetectable expression of endogenous GFP-tagged KIN-29 (Fig. 1F).
To determine if KIN-29’s regulation of sleep has a developmental component, we used two paradigms. First, we cultivated young adult animals for four hours on agar containing auxin (2 mM). We then administered a UVC treatment (1500 J/m2) and recorded movement quiescence of animals in the continued presence of auxin. Four hours in the presence of auxin at the young adult stage was sufficient to reduce GFP-tagged KIN-29 expression (Fig. 1F), and was able to reduce SIS to a similar extent as that seen in animals cultivated on auxin their whole life (Fig. 1F-G). Second, to restore KIN-29 function to adult animals, we cultivated KIN-29 AID transgenic animals in the presence of auxin since hatching, but then removed them from auxin-containing plates at the L4 stage. We waited twelve hours before assessing SIS using UVC-irradiation treatment. These animals showed significantly increased movement quiescence in comparison to animals that were cultured their whole life on auxin, although these quiescence levels were lower than the quiescence phenotype of wt animals (Fig. 1H). Consistent with these intermediate behavioral results, expression of GFP-tagged KIN-29 in these animals was intermediate between that of animals never exposed to auxin, and that of animals exposed to auxin throughout life (Fig. 1F). Our results are consistent with previous observations that even after 24 hours off of auxin, expression of a DEGRON-tagged protein may not fully normalize (Zhang et al. 2015).
In summary, our results demonstrate that KIN-29’s function in SIS is not developmental. Rather, these findings suggest that kin-29 functions at the time of sleep. These results fit with our proposed model of kin-29 acting as a real-time energy sensor in sensory neurons to promote sleep (Grubbs et al. 2020). Based on this analysis, we propose that the sleep-regulatory effects of SIK3 in mammals (Funato et al. 2016) are also unlikely to be developmental.
Methods
Request a detailed protocolWorm maintenance and strains
Worms were cultivated on the agar surface of 5.5 cm diameter Petri dishes filled with 11.5 mL of NGM agar. They were fed the Escherichia coli strain DA837 (Davis et al. 1995) and grown at 20°C. All experiments were performed on hermaphrodites. The wild-type strain used was N2, variety Bristol (Brenner 1974). Auxin experiments were carried out as described in (Zhang et al. 2015) using 2 mM auxin (Alfa Aesar #A10556). Strains available from the CGC are N2 and CA1200. Strains available from our lab are PHX1777 kin-29(syb1777[kin-29::degron::gfp]) and NQ1295 (ieSi57[Peft-3::tir1::mRuby]; kin-29(syb1777[kin-29::degron::gfp]).
CRISPR/Cas9 genome editing and crosses
CRISPR/Cas9-mediated genome editing was performed by SunyBiotech (China). The oligonucleotide sequence used for PCR and Sequencing were (Sense) 5’CATTGTGTGCACATTACCGT and (Antisense) 5’AATCAGCTAGCACAGGCTCT. To generate the strain NQ1295, PHX1777 males were crossed with CA1200 hermaphrodites, with the red and green fluorescent phenotypes followed using a Leica SP8 confocal microscope. We found a regular epifluorescence compound microscope not sufficiently sensitive to detect the fluorescence in these strains, which was very dim.
Ultraviolet light exposure and movement quiescence recordings
Movement quiescence was quantified using machine vision with images recorded using the 48-well WorMotel (Churgin et al. 2019). Briefly, animals were selected as L4 larvae 12-16 hours prior to behavioral recording. All 48 wells were filled with NGM agar with or without 2 mM auxin, which was diluted from a 400 mM stock as described in (Zhang et al. 2015), and a thin layer of bacteria was spread over the agar surface using a platinum wire. Animals were loaded individually as adults into WorMotel wells and then treated using a Spectrolinker XL-1000 crosslinker with 1500 J/m2 of UVC (254 nm) radiation. The WorMotel was placed under a DMK 23GP031 camera (The Imaging Source) for recording. Images were captured every 10 seconds for 8 hours. Images collected of the animals on the WorMotel were analyzed with custom MatLab scripts (Churgin et al. 2019). The quantification of movement was determined via pixel subtraction and an animal was considered quiescent if there was no pixel change between images.
Confocal imaging
Animals were placed in a 1ml droplet of M9 buffer and immobilized on a 2% agarose pad containing 25mM levamisole. Images were taken using a Leica SP8 confocal microscope as Z-stacks with 0.76 microns between images in a stack and a total of 36 images per condition taken. The powers of the 488 nm and 552 nm lasers were set to 2.5% and 2% respectively. Z-projection was performed in FIJI (Version 2.0.0).
Acknowledgments
We would like to thank SunyBiotech for generation of the kin-29::degron::GFP tagged (PHX1777) strain.
References
Funding
This work was supported by the National Institutes of Health grant R01NS107969. The CA1200 strain was provided by the Caenorhabditis Genetics Center, which is funded by the NIH office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
Cheryl Van BuskirkHistory
Received: April 28, 2020Revision received: May 6, 2020
Accepted: May 7, 2020
Published: May 7, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Grubbs, JJ; van der Linden, AM; Raizen, DM (2020). Regulation of sleep by KIN-29 is not developmental. microPublication Biology. 10.17912/micropub.biology.000247.Download: RIS BibTeX