Description
Mutator foci are perinuclear granules in the germline of Caenorhabditis elegans that are required for the amplification of 22G-small interfering RNAs (siRNAs) (Phillips et al., 2012). These mutator-dependent siRNAs act downstream of primary endogenous and exogenous siRNA pathways and are necessary for robust and heritable silencing (Pak et al., 2007; Sijen et al., 2007; Gu et al., 2009; Gent et al., 2010; Vasale et al., 2010; Phillips et al., 2012). There are numerous factors that have been identified that localize to Mutator foci and are required for mutator-dependent siRNA biogenesis. These mutator-class proteins include the core component of Mutator foci MUT-16, the nucleotidyl transferase MUT-2, the 3’-5’ exonuclease MUT-7, the DEAD-box RNA helicases MUT-14 and SMUT-1, the Zc3h12a-like ribonucleases RDE-8, NYN-1, and NYN-2, and two proteins of unknown function, MUT-15 and RDE-2 (Ketting et al., 1999; Tijsterman et al., 2002; Vastenhouw et al., 2003; Chen et al., 2005; Tops et al., 2005; Phillips et al., 2012; Phillips et al., 2014; Tsai et al., 2015). Additionally, the RNA-dependent RNA polymerase RRF-1 localizes to Mutator foci but is redundant with EGO-1 for mutator-dependent siRNA biogenesis (Phillips et al., 2012; Gu et al., 2009). It was previously shown that mutations in mutator-class genes are sterile at elevated temperature (Ketting et al., 1999; Zhang et al., 2011; Rogers and Phillips, 2020). Recently, we performed a brood size assay using wild-type and mut-16 hermaphrodites cultured at 20°C. We found that compared to wild-type animals, mut-16 mutant animals lay fewer eggs (56% fewer eggs laid compared to wild-type animals), and of those eggs, fewer mut-16 mutant eggs hatch (81% of mut-16 mutant eggs hatch compared to wild-type, where 100% of the eggs hatch) (Rogers and Phillips, 2020). Furthermore, 100% of wild-type larvae successfully mature to adulthood, whereas only 85% of mut-16 mutant larvae mature to adulthood (Rogers and Phillips, 2020). The reduced hatching rates and larval arrest of mut-16 mutant animals had not been previously reported.
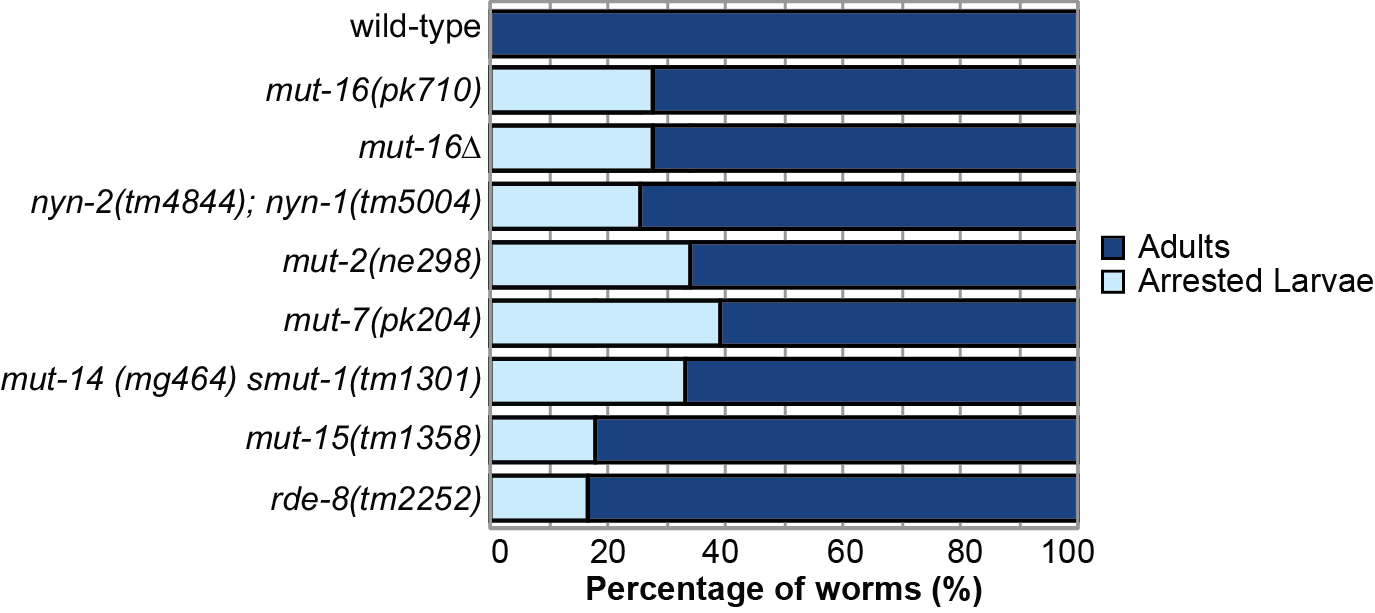
Because one phenotype of mutants of the mutator-class genes is hopping of transposable elements, and thus they can exhibit spontaneous mutations (Ketting et al., 1999), in this work we first sought to test whether the larval arrest phenotype is found in other mut-16 mutant alleles and not due to a background mutation in the mut-16(pk710) strain. We performed a larval arrest assay in which we counted the total number of individuals that mature to adulthood or arrest as larvae for wild-type (N2) and mut-16 mutants. We used two mut-16 alleles, mut-16(pk710), the same allele as the original assay which carries an early stop codon, and mut-16(cmp185) (referred to here as mut-16Δ), an in-frame 560 amino acid deletion (Uebel et al., 2018). One thousand L1 stage animals of each strain were plated at 20°C. After seventy-two hours, the developmental stage of the individuals was assessed. We found that 28% of both mut-16Δ and mut-16(pk710) mutant individuals arrested as larvae compared to 0% of wild-type individuals (Figure 1).
To determine whether larval arrest is a common phenotype amongst other mutator mutants, we counted the larval arrest rate for six additional mutator-class mutants. We observed that a portion of the L1 stage animals from each mutator-class mutant arrested at either the L1 or L2 stage – nyn-1(tm5004); nyn-2(tm4844) mutants (26% arrest), mut-2(ne298) mutants (34% arrest), mut-7(pk204) mutants (39% arrest), mut-14(mg464) smut-1(tm1301) mutants (33% arrest), mut-15(tm1358) mutants (18% arrest), and rde-8(tm2252) mutants (16% arrest) (Figure 1). These data indicate that larval arrest is a low penetrance phenotype found in between 16% and 39% of mutant individuals, and that mutator-class proteins, such as MUT-16, play an important role in ensuring the development of C. elegans.
While elevated temperature triggers sterility in mutator-class mutants, here we show that mutator-class mutants also exhibit a larval arrest phenotype at permissive temperature. Larval arrest can occur in C. elegans for many reasons, including but not limited to, stressful conditions such as starvation – which could occur due to a lack of food (Johnson et al., 1984), the inability to consume food or perform pharyngeal pumping (Fay et al., 2003; Furuya et al., 2005; Mango, 2007), or the inability to absorb nutrients in the gut (Thieringer et al., 2003) – or mis-regulation of cell cycle components (Boxem et al., 1999; van den Heuvel, 2005), proteasome components (Takahashi et al., 2002), or other pathways that affect development. The individuals assayed in our experiments were not grown under stressful conditions or on densely populated plates. Thus, the underlying cause of larval arrest in mutator-class mutants could be due to the inability of the animals to consume food, absorb nutrients, or due to mis-regulation of factors necessary for proper development of C. elegans. Previously, we showed that mut-16 has a maternal and paternal effect on sterility when animals are raised at elevated temperature (25°C) (Rogers and Phillips, 2020). Thus, the low penetrance larval arrest phenotype of mut-16 mutants could arise from maternal effects, paternal effects, zygotic effects, or a combination. Further experiments will be needed to determine the underlying cause of larval arrest in mutator-class mutants. It is interesting to note that when mut-16 mutant larvae are synchronized by starvation 28% arrest as larvae, whereas when we previously performed a brood size assay, where no L1 starvation occurred, 15% of mut-16 mutant larvae arrest (Rogers and Phillips, 2020). This difference suggests that, while the larval arrest phenotype of mutator mutants can occur either when larvae hatch from eggs in the presence of food or when starved as L1s, starvation may exacerbate the arrest phenotype. Taken together with the reduced egg laying of mut-16 mutants (Rogers and Phillips, 2020), these findings suggest that MUT-16, and other mutator-class proteins, play key roles in both maintaining fertility and promoting development in C. elegans.
Methods
Request a detailed protocolC. elegans strains. All animals were grown at 20°C according to standard conditions (Brenner 1974). All strains are in the wild-type (N2) background and have been outcrossed at least four times.
Larval arrest assay. Worms were synchronized by bleaching and were then plated on NGM plates, 20 L1 stage animals per plate and one thousand individuals per genotype. After 72 hours at 20°C, the number of individuals that reached adulthood or arrested as L1-L2s were counted.
Reagents
N2 – wild-type.
NL1810 – mut-16(pk710) I.
GR1747 – mut-15(tm1358) V.
GR1948 – mut-14(mg464) smut-1(tm1301) V.
WM30 – mut-2(ne298) I.
NL1820 – mut-7(pk204) III.
FX2252 – rde-8(tm2252) IV.
USC880 – nyn-2(tm4844) I; nyn-1(tm5004) IV.
USC1148 – mut-2(cmp42[(mut-2::gfp::3xFLAG)]) mut-16(cmp185[mut-16ΔE-K::mCherry::2xHA]) I.
We used USC1148 (referred to here as mut-16Δ) for the mut-16 deletion allele. It should be noted that USC1148 contains MUT-2::GFP::3xFLAG, which does not affect the function of MUT-2.
Acknowledgments
We would like to thank members of the Phillips lab for discussion.
References
Funding
This work was supported in part by a Basil O'Connor Starter Scholar Research Award from the March of Dimes Foundation [5-FY17-38 to CMP] and the National Institute of Heath [R35 GM119656 to CMP]. CMP is a Pew Scholar in the Biomedical Sciences supported by the Pew Charitable Trusts.
Reviewed By
Rob DowenHistory
Received: April 7, 2020Revision received: May 15, 2020
Accepted: May 18, 2020
Published: May 19, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Rogers, AK; Phillips, CM (2020). Disruption of the mutator complex triggers a low penetrance larval arrest phenotype. microPublication Biology. 10.17912/micropub.biology.000252.Download: RIS BibTeX