Description
Neurodegenerative diseases caused by short expansive repeats like the (CAG) in Huntington’s disease (Orr 2012) or the (GGGGCC) repeat in C9orf72-associated Amyotrophic lateral sclerosis (ALS)/Frontotemporal dementia (FTD) (DeJesus-Hernandez et al. 2011) undergo an unusual type of translation called repeat associated non-AUG-dependent (RAN) translation (Cleary and Ranum 2014). Interestingly, RAN translation occurs without an AUG start codon (Cleary and Ranum 2014). This allows for the (GGGGCC) repeat mutation to be translated, even though it is located in the intron between exon 1 and exon 2 of the C9orf72 gene, which would normally be spliced out and degraded (DeJesus-Hernandez et al. 2011). Translation of the repeat occurs in all 3 reading frames, leading to the production of three distinct dipeptide repeat proteins (DPRs). RAN translation begins within the (GGGGCC) repeat, but the exact translation initiation site remains unclear. However, RAN translation does not stop at the end of the repeat and will continue to translate the intronic sequence until it reaches a stop codon. This means that each of the distinct DPRs will be fused to peptides encoded in the downstream intron sequence. Because the DPRs are derived from intron sequence that is spliced out of the mature C9orf72 mRNA, none of these intron-derived DPR fusion peptides are incorporated into the ‘normal’ C9orf72 protein. While it is known that the DPR fusion peptides are made in patients, the precise sequences of the DPR fusion peptides that they produce is not currently known. Therefore, questions about where precisely RAN translation initiates, how many repeats are produced, and whether the number of repeats produced are uniform or heterogenous remain important but unresolved questions.
There is also a C9orf72 antisense transcript, which contains the complementary repeat sequence (GGCCCC). This antisense transcript also undergoes RAN translation to produce another three DPRs (Zu et al. 2013). Therefore, a single DNA repeat expansion in one gene gives rise to six distinct DPRs. These DPRs form p62 positive/pTDP-43 negative inclusions that are distinct hallmarks of C9orf72-associated ALS/FTD (Cleary and Ranum 2014). Our laboratory as well as others have shown that two of these DPRs, proline-arginine (PR) and glycine-arginine (GR) are highly toxic (Kwon et al. 2014; Wen et al. 2014; Rudich et al. 2017), however the mechanisms of toxicity are poorly defined.
In order to study the mechanisms that cause C9orf72-associated ALS/FTD PR and GR toxicity, we utilized the Caenorhabditis elegans model system. With short lifespans (3-4 weeks), a conserved neuromuscular system, and a genome that encodes ~20,000 genes with many conserved human homologs, the C. elegans model system is highly relevant for the study of aging and age-related diseases like ALS (Olsen et al. 2006). To study how PR and GR cause toxicity in C. elegans, we created animals expressing codon-optimized (PR)50-GFP and (GR)50-GFP (Rudich et al. 2017). With this approach, we are able to observe the effects of a single DPR at a time, without additional contributions from either the loss of the C9orf72 gene expression, introduction of the G4C2 repeat containing RNA, or the other five RAN translated DPRs. Therefore, this is a pure DPR model. Our laboratory has previously shown (PR)50 and (GR)50 to be toxic by causing a decrease in motility (paralysis) and arrested growth, when expressed in muscle (Rudich et al. 2017). Nuclear localization of these two DPRs was also discovered to be necessary and sufficient for toxicity in C. elegans (Rudich et al. 2017).
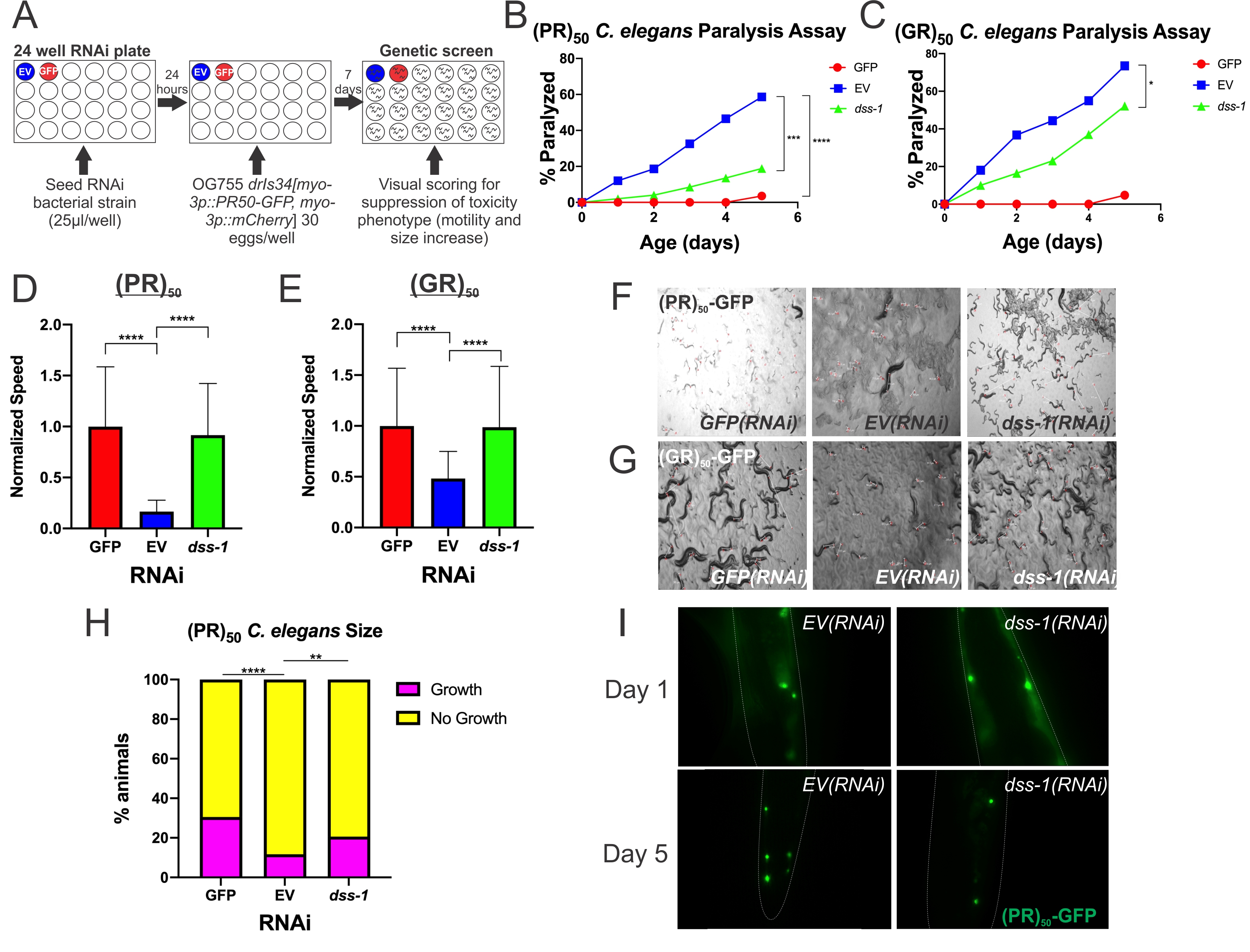
Using a transgenic C. elegans (PR)50 line, we screened RNAi feeding clones targeting 1,691 genes that are unique to the ORFeome RNAi library and not found in the MRC/Ahringer RNAi library. GFP(RNAi) and EV(RNAi) functioned as positive and negative controls respectively (Figure 1A). After two rescreens of the initial hits from the 1,691 genes screened, we identified two RNAi knockdowns that were able to suppress the loss of motility and arrest in growth caused by (PR)50. These two hits were subsequently screened six additional times, all of which suppressed (PR)50 growth arrest and paralysis. Both RNAi clones were sequenced and the sequences were aligned to the C. elegans genome using BLAST to identify the affected genes. The two genes uncovered in the screen were lin-54 and dss-1. In most assays, dss-1(RNAi) had a much stronger toxicity suppression phenotype than lin-54(RNAi) and rescued motility back to near wildtype levels. Because the dss-1(RNAi) phenotype was robust, additional mechanistic studies were performed to understand how dss-1 contributes to DPR toxicity.
One possible mechanism for the observed suppression was a decrease in the activity of the promoter (myo-3p) controlling (PR)50 gene expression. In C. elegans RNAi screens utilizing transgene-based phenotypes (such as the transgenic PR expression utilized here), one known class of suppressors include genes whose inhibition globally suppresses transgene expression (Fischer et al. 2013). Large object flow cytometry (COPAS sorting) was used to quantitatively measure the expression level of a fluorescent reporter (Red fluorescent protein, RFP) derived from the PR transgene which utilizes the same myo-3 promoter (Rudich et al. 2017). Normalized RFP levels (RFP/TOF) were not significantly different between EV(RNAi) and dss-1(RNAi) ((EV(RNAi) – .04615 +/- .03731; N = 30; dss-1(RNAi) – .05432 +/- .08057; N=55; ANOVA with post-hoc test, p > 0.9999). Therefore, dss-1(RNAi) does not act via transgene suppression.
When (PR)50 is expressed post-developmentally in adult C. elegans, it causes an age-dependent paralysis phenotype. GFP(RNAi) fully suppresses age-dependent paralysis and restores motility, strongly suggesting that this phenotype is due to the expression of the (PR)50 protein. Using this paralysis assay, we examined the effect that dss-1(RNAi) had on age-dependent paralysis of C. elegans expressing (PR)50. Two independent paralysis assays showed that animals on dss-1(RNAi) exhibited reduced paralysis compared to the negative control EV(RNAi) (Figure 1B). In addition to these phenotypes, the other toxic DPR made via the C9orf72 repeat expansion mutation, (GR)50, showed similar results (Figure 1C). This decrease in C. elegans age-dependent paralysis following dss-1(RNAi) shows that dss-1 normally functions to facilitate (PR)50 and (GR)50 induced age-dependent paralysis.
When (PR)50 is expressed developmentally in muscle, it causes strong larval paralysis. Larval paralysis is completely suppressed by GFP(RNAi), strongly suggesting that this phenotype is due to the expression of the (PR)50-GFP protein. In order to examine the effect that (PR)50 has on the developmental paralysis phenotype, we developed a novel method called video speed analysis (VSA) (Rudich et al. 2020). Two independent VSA trials showed increased motility in (PR)50 expressing animals on dss-1(RNAi) compared to EV(RNAi) (Figure 1D, F). (GR)50, showed similar results (Figure 1E, G). These results display that dss-1 normally functions to help facilitate PR and GR induced developmental paralysis.
When (PR)50 is expressed developmentally in muscle, it causes an arrest in the growth of C. elegans along with a decrease in motility. This results in the accumulation of short larvae and a depletion of longer adult worms. Feeding animals GFP(RNAi) completely suppresses this growth arrest, strongly suggesting this phenotype is due to the (PR)50 protein. In order to quantify the effect of (PR)50 +/- dss-1(RNAi) on C. elegans size, C. elegans length was measured via COPAS biosorting on GFP(RNAi), EV(RNAi), and dss-1(RNAi) (Figure 1H). Setup of C. elegans for sorting followed the same methodology and time frame as the RNAi genetic screen. ‘Growth’ or ‘No Growth’ thresholds were empirically determined based on the sizes observed in the positive (GFP(RNAi)) and negative (EV(RNAi)) controls. Worms above this threshold were labeled as ‘Growth’ and worms below this threshold were labeled as ‘No Growth.’ Results from this experiment showed that C. elegans increased in size on dss-1(RNAi) compared to EV(RNAi), showing that dss-1(RNAi) suppressed the growth arrest caused by (PR)50 (Figure 1H). This result shows that dss-1 normally functions to help facilitate (PR)50-induced suppression of growth.
As we previously discovered in our laboratory, nuclear localization is required for (PR)50 and (GR)50 toxicity in C. elegans (Rudich et al. 2017). Therefore, dss-1(RNAi) could suppress (PR)50 toxicity either because it prevents (PR)50 localization to the nucleus or because it inhibits relevant toxicity pathways downstream of nuclear (PR)50. To distinguish between these possibilities, we used fluorescence microscopy to determine if (PR)50-GFP localization is altered by dss-1(RNAi) (Figure 1I). In dss-1(RNAi) animals, PR nuclear localization was unaffected. Therefore, dss-1 is not required for PR nuclear localization but rather facilitates the (PR)50 toxicity mechanism(s). The possible mechanisms through which dss-1 functions in this pathway to cause toxicity are unclear and will be a point of future research.
Through these results, we have shown that dss-1 is required for C9orf72-associated ALS/FTD. dss-1 (Deleted in Split hand/Split foot protein 1) is a small conserved (human ortholog Sem1) multi-functional nuclear protein involved in multiple different cellular processes (Pispa et al. 2008). dss-1 is part of the 26S proteasome, helps assemble the 19S subunits of the 26S proteasome, and binds ubiquitin (Kragelund et al. 2016). While it is still unclear what the ubiquitin binding role(s) of dss-1 are in the 26S proteasome, it is hypothesized that it may act as a ubiquitin receptor domain within the proteasome (Paraskevopoulos et al. 2014). In addition, dss-1 is also part of the Sac3-Thp1, or TREX-2, complex which is involved in mRNA export as well as the Csn12-Thp3 complex which is involved in RNA splicing in the nucleus (Wilmes et al. 2008). Both the proteasome and mRNA export/splicing have been previously linked to C9orf72-associated DPR toxicity (Boeynaems et al. 2017, Gupta et al. 2017, Kramer et al. 2018, Kwon et al. 2014, Lee et al. 2016).
dss-1 is characterized as an intrinsically disordered protein due to its lack of defined structure and its ability to have multiple protein conformations (Kragelund et al. 2016). Interestingly, it has been shown that many intrinsically disordered proteins promote liquid-liquid phase separation (LLPS), an important organizing mechanism for intracellular compartmentalization of some proteins (Alberti 2017). The disruption of LLPS has been identified in multiple neuropathologies, including C9orf72-associated ALS/FTD (Boeynaems et al. 2017). Toxic DPRs PR and GR have been shown to be able to phase separate, which can disrupt LLPS of nucleoli, the nuclear pore complex and stress granules (Lee et al. 2016; Boeynaems et al. 2017). These stress granules contain important RNA binding proteins like TDP-43 and FUS (Boeynaems et al. 2017). PR and GR have been shown to disrupt these stress granules by causing liquid-to-solid maturation (Boeynaems et al. 2017). This liquid-to-solid formation is a hallmark of neurodegenerative pathogenesis and triggers an acceleration of ALS phenotypes (Patel et al. 2015). Together, these observations suggest that dss-1 may play an important role in LLPS pathology that influences ALS/FTD toxicity.
A recent study shows that the proteasome, including the 26S regulatory subunit with which dss-1 interacts, also undergoes ubiquitin-dependent LLPS in the nucleus (Yasuda et al. 2020), suggesting a plausible connection among dss-1, C9orf72-associated PR and GR liquid-liquid phase separation (LLPS), and the proteasome. These inter-connections include: PR and GR mediated inhibition of ubiquinated substrates (Gupta et al. 2017); CRISPR-Cas9 screens in K562 cells and primary mouse neurons showing proteasome subunits as genetic modifiers of PR and GR (Kramer et al. 2018); and C9orf72-associated impairment of TDP-43 degradation by the proteasome (Lee et al. 2019). However, dss-1 may offer a new link in regard to C9orf72-associated ALS/FTD and the proteasome. One potential hypothesis is that dss-1 helps facilitate LLPS disruption of the proteasome by C9orf72-associated toxic DPRs PR and GR. Since ubiquitinated substrates are required for liquid droplet formation of the proteasome (Yasuda et al. 2020), the role of dss-1 as a ubiquitin receptor in the proteasome (Paraskevopoulos et al. 2014) may help in the formation of liquid droplets. Loss of dss-1 could be protective because there is less LLPS of the proteasome due to a lack of recruitment of ubiquinated substrates. Therefore, with a reduction in LLPS, the toxic DPRs can no longer cause liquid-to-solid maturation which has been shown to be harmful (Patel et al. 2015). Future experimentation testing this hypothesis, along with investigation into the other known functions of dss-1 in mRNA nuclear export and mRNA splicing, may lead to insights into a conserved mechanism by which dss-1 is required for PR and GR toxicity.
In conclusion, this study highlights dss-1 as a key component of the C9orf72-associated ALS/FTD genetic pathway and suggests dss-1 as a potential therapeutic target for treatment of ALS/FTD. This study has shown that dss-1 inhibition is able to rescue developmental and post-developmental motility as well as developmental size defects in C. elegans expressing the toxic (PR)50 and (GR)50 dipeptide repeat proteins. Future studies in mice and human cell culture are needed to evaluate the potential efficacy of dss-1 antagonists as a therapeutic option for treating C9orf72-associated ALS/FTD patients.
Reagents
C. elegans Strains: OG755 +/+; drIs34 [myo-3p::(PR)50-GFP; myo-3p::mCherry] X and OG736 +/+; drIs28 [myo-3p::(GR)50-GFP; myo3p::mCherry] (integration site unmapped); C. elegans ORFeome RNAi library (Dharmacon/Horizon Discovery, Cambridge, UK), dss-1 RNAi clone ORF ID – Y119D3B.15
References
Funding
R21NS096319, R21NS094921
Reviewed By
Alex ParkerHistory
Received: April 22, 2020Revision received: May 15, 2020
Accepted: May 27, 2020
Published: June 2, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Puleo, N; Lamitina, T (2020). The conserved multi-functional nuclear protein dss-1/Sem1 is required for C9orf72-associated ALS/FTD dipeptide toxicity. microPublication Biology. 10.17912/micropub.biology.000262.Download: RIS BibTeX