Baylor University Department of Biology
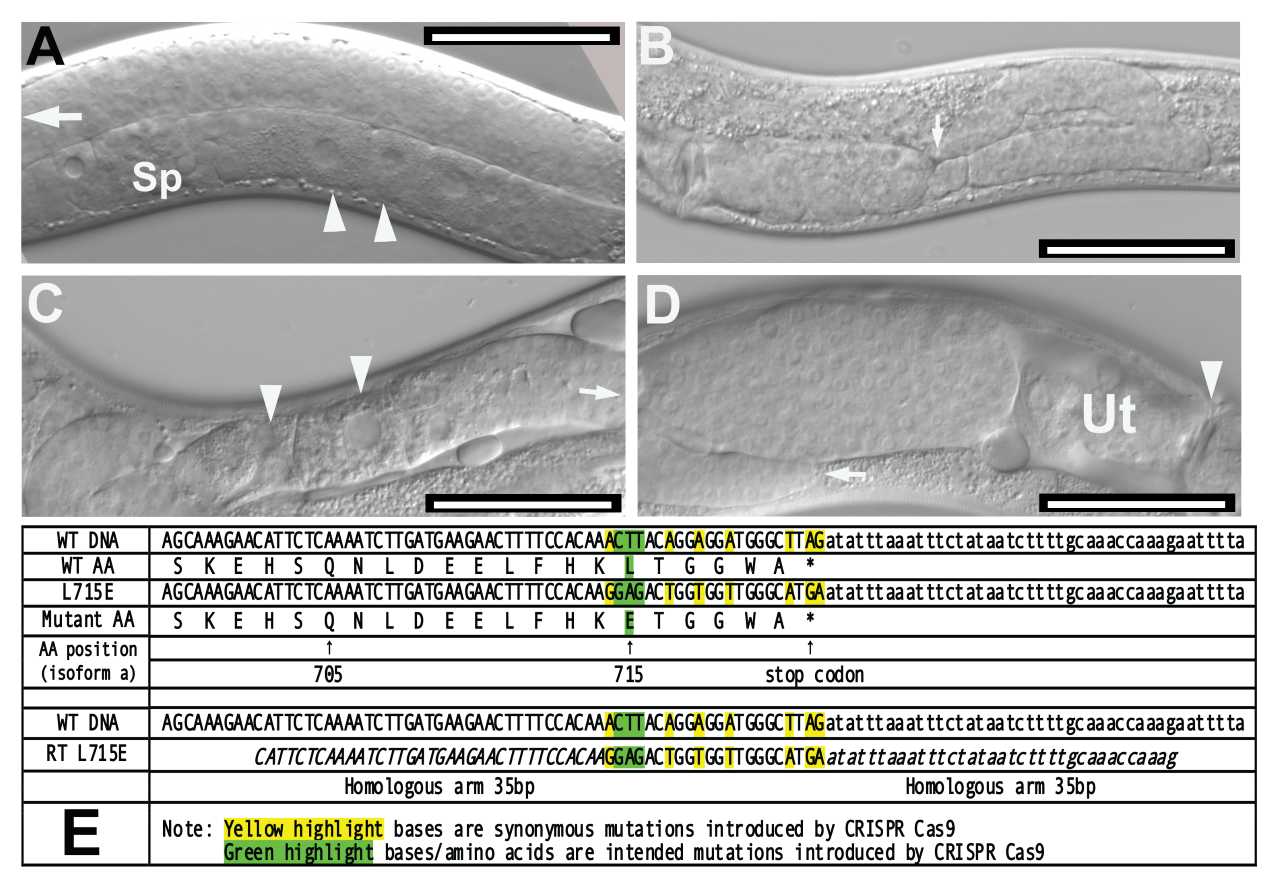
A. N2 Caenorhabditis elegans displaying normal gonad. White arrowheads indicate developing oocytes in the ventral side of the organism (proximal gonad). The distal gonad is filled with undifferentiated germ cell nuclei, white arrow. Sp: spermatheca. Bar = 50 μm.
B. unc112(kq715) mutant displaying distal tip cell migration defects (DTC Mig). White arrow indicates abnormal “dipping” of the distal arm. Bar = 50 μm.
C. unc-112(kq715) mutant displaying ectopic oocytes. White arrowheads indicate meiotic oocytes developing in the distal gonad arm. Arrow indicates the direction of DTC. Bar = 50 μm.
D. unc-112(kq715) mutant displaying abnormal bulging gonad filled with undifferentiated germ cells between label D and uterus (Ut). Bar = 50 μm.
E. Comparison of wild-type unc-112 amino acid sequence and the mutant sequence including the repair template sequence and synonymous mutation schemes. All sequences are written in the 5’ to 3’ direction. *WT: wild-type. **AA: amino acid.
Description
The unc-112 gene in Caenorhabditis elegans encodes a 720-amino acid protein (UNC-112) that colocalizes with integrin and perlecan in cell to ECM adhesion structures and is a component of dense bodies and M-lines (Rogalski et al., 2000). The UNC-112 protein is homologous to the human protein Kindlin-1. Kindlin-1 in humans is implicated in Kindler syndrome, a skin fragility disorder (Siegel et al., 2003). The UNC-112 protein shares a short region of homology (roughly 200 amino acids) with talin and other FERM superfamily proteins that may play a key role in plasma membrane attachment (Rogalski et al., 2000). Previous studies have discovered that UNC-112 is essential for the organization and localization of PAT-3/β-integrin in body wall muscle cells (Rogalski et al., 2000). Null mutations in unc-112 lead to twofold stage arrest during embryogenesis, abnormal body wall muscle, and exhibit the Pat (paralyzed, arrested elongation at twofold) phenotype (Williams and Waterston, 1994). In this study, a novel mutant allele of C. elegans unc-112 was generated using CRISPR-Cas9 gene editing. In this mutant allele, leucine (L) was replaced with glutamate (E) near the C-terminal end of UNC-112. In a biochemical study, the homologous mutation abolished binding of kindlin2 to the membrane distal “phospho-tyrosine” motif of β1 integrin tails (Li et al., 2017). Mutant worms were examined for behavioral and morphological abnormalities. Mutants displayed no notable behavioral abnormalities in a thrashing assay compared to N2 wild-type worms (p=0.5485 according to the Mann-Whitney U Test). However, 35.6% of unc-112 mutants (n=177) were observed to exhibit defective distal tip cell migration (DTC Mig). Mutants displaying this abnormal DTC Mig phenotype had dipping distal gonad arms (Figure 1B), bulging proximal gonads filled with germ cell nuclei (Figure 1D), and oocyte-like nuclei in the distal gonad (ectopic oocytes, Figure 1C). Previous research has shown that integrin-ECM interactions play important roles in directing DTC migration and gonad morphogenesis, and the DTC Mig phenotype observed could be the result of mutant unc-112 disrupting key integrin functions in the somatic gonad (Lee et al., 2001)
Methods
Request a detailed protocolIn order to induce the desired mutation to unc-112, the CRISPR guide RNA Selection Tool (http://genome.sfu.ca/crispr/) was used to identify CRISPR target sites within the gene. The mutant line created, unc-112(kq715), contains a mutation from Leucine to Glutamate at amino acid number 715 near the C-terminal end of UNC-112. Custom template DNA (Temp-4UNC112L715E, Figure E), custom crRNA (UNC112STOP), tracrRNA (cat. no. 1073190), and Alt-R Cas9 nuclease (cat. no. 1081058) were annealed at room temperature (Paix et al.., 2015) and micro-injected into the syncytial gonad arms of N2 wild-type worms (P0) (Mello et al., 1991). F1 dumpy (Dpy), co-CRISPR phenotype, worms were isolated and PCR screened for the presence of the desired mutation. The offspring from F1 heterozygous mutants were then PCR screened to isolate homozygous mutants (F2). The isolated mutant line was backcrossed (2x) to N2 and were then studied for phenotype and behavioral characterization. In order to identify DTC Mig phenotypes, N2 and unc-112(kq715) worms were self-fertilized and screened for aforementioned distal tip cell abnormalities. Briefly, animals were mounted and observed on a Nikon Eclipse Ni-U compound microscope and images were captured using NIS Elements software (version 5.02). The unc-112 mutant animals with DTC migration defects were examined with Nomarski optics. The morphology of U-shaped gonad arms was observed in gonad arms at the L4 or young adult stage of hermaphrodites. The percentage of abnormal gonad arms, displaying no looping back, zigzag distal arms, or extra turns were calculated (Lee and Cram, 2009). The thrashing assay was performed by counting the number of body bends over a 10 second period when suspended in a 10 μl drop of M9 buffer (Lee et al., 2005). A Mann-Whitney U Test was then run to confirm statistical significance of thrashing assay results
crRNA sequence
UNC112STOP: CCACAAACTTACAGGAGGAT
PCR Primers
UNC112L715SEQF: AACTTGGTGCTGACAGGAAGG
UNC112L715SEQR: GCGAGATATGCGAAACGTTGA
UNC112L715WTR: ATCTAAGCCCATCCTCCTGTAAGT
UNC112L715ER: TCATGCCCAACCACCAGTCTCC
Reagents
BU0715 unc-112(kq715) is available upon request.
Acknowledgments
This study was conducted during the course of BIO 4108 at Baylor University.
References
Funding
Funding for BIO 4108 is provided by Baylor University.
Reviewed By
Kiyoji NishiwakiHistory
Received: May 22, 2020Revision received: June 3, 2020
Accepted: June 3, 2020
Published: June 8, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Park, A; Qiu, Z; Bumm, J; Lee, M (2020). A novel mutation in unc-112/kindlin locus causes distal tip cell migration defects. microPublication Biology. 10.17912/micropub.biology.000265.Download: RIS BibTeX