Department of Biology, University of Iowa, Iowa City IA
Description
In C. elegans, RNAi is performed by delivering double-stranded RNA (dsRNA) to the worm via feeding (Timmons and Fire 1998), injection (Fire et al. 1998) or soaking (Tabara, Grishok, and Mello 1998). In feeding RNAi, sequence of a target gene is placed between two oppositely oriented promoter sequences active in E. coli (e.g. T7 in the pL4440 vector (Timmons and Fire 1998)). Subsequent T7 polymerase expression in E. coli provides sense and anti-sense transcription of the target gene sequence, generating dsRNA. The bacteria producing the dsRNA is consumed by the worm and the dsRNA distributed to C. elegans cells (Timmons and Fire 1998), resulting in efficient target gene knockdown.
When performing RNAi by feeding, one can achieve a range of knockdown phenocopies by diluting the bacteria producing dsRNA targeting the gene of interest with similarly prepared bacteria producing dsRNA from empty pL4440 vector. The ability to dilute the level of knockdown of a target gene is useful when undiluted RNAi causes developmental defects, or if a partial loss of function is preferred over a full gene knockdown. While mixing with empty vector bacteria sufficiently dilutes the target gene bacteria fed to the worm, it may not be sufficient to dilute the silencing RNAs loaded into RNAi riboproteins. Specifically, an “empty” vector plasmid produces a short amount of dsRNA consisting of only 185bp of polylinker sequence. Thus, there will be few dsRNA segments in worm cells after empty vector RNAi feeding. Therefore, dilution of a desired target gene RNAi vector, which often produces closer to 1000bp of dsRNA, with empty vector plasmid may result in a preponderance of RNAi riboproteins loaded with target gene dsRNA, minimizing the dilution effect. Here, we asked if dilution with a plasmid generating a longer dsRNA than empty vector would be more effective at diluting the target gene mRNAs leading to a partial loss of function phenotype.
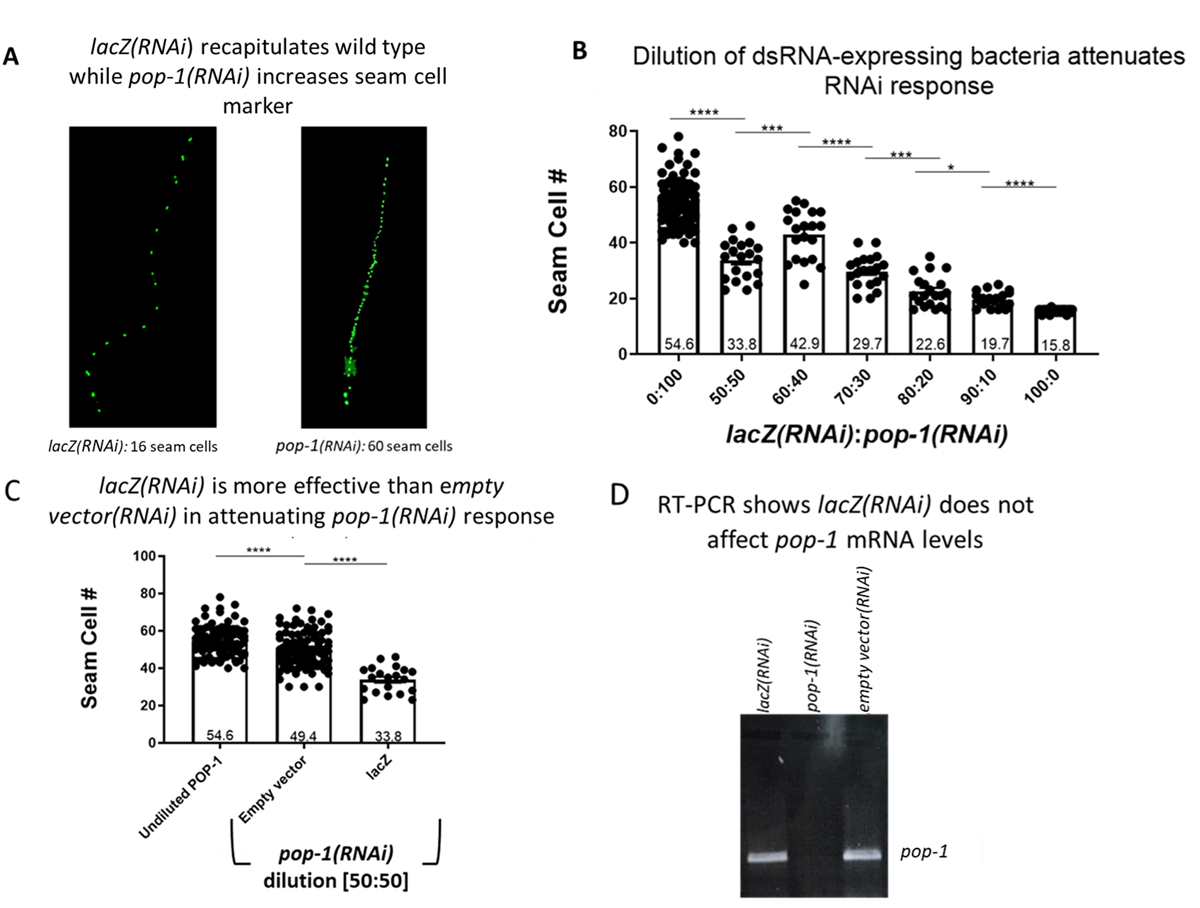
To begin, we identified a gene that displayed allele-dependent variations in expressivity. We decided to evaluate the effect of pop-1/TCF knockdown on seam cell output since knockdown of POP-1-mediated transcriptional repression via feeding RNAi is known to result in an increase from the wild-type number of 16 seam cells to as many as 60 seam cells (Banerjee et al. 2010). The seam cells undergo several rounds of Wnt-signaled asymmetric cell divisions where low nuclear POP-1/TCF in the posterior daughter confers seam cell fate while higher POP-1/TCF in the anterior daughter results in hypodermal cell fate (Baldwin and Phillips 2018; Baldwin, Clemons, and Phillips 2016; Lam and Phillips 2017; Lin, Hill, and Priess 1998; Takeshita and Sawa 2005). To utilize seam cell number as an output, we performed our experiments in the strain JR667 which marks the seam cell fate with GFP from the wIs51 transgene (Figure 1A). To determine if a RNAi vector encoding longer dsRNA sequences was a more effective dilutor than the pL4440 empty vector, we inserted a portion of the E. coli lacZ gene, which is absent from the C. elegans genome, into pL4440. The resulting lacZ(RNAi) vector produces 1255bp of dsRNA (1070bp of lacZ plus 185bp polylinker), which is a similar length to our pop-1(RNAi) vector that generates 1292bp dsRNA (1107bp of pop-1 cDNA plus 185bp polylinker).
In order to find an RNAi dose that results in an intermediate pop-1 phenocopy that can be used as a baseline for different dsRNA dilution protocols, we showed that bacteria mixing of pop-1(RNAi) with lacZ(RNAi) at different ratios corresponded to different increases in seam cell output (Figure 1B). Generally, highly diluted pop-1(RNAi) bacterial cultures corresponded to a mild increase in seam cells (e.g. 19.7 seam cells at the 90:10 dilution of lacZ(RNAi):pop-1(RNAi)) whereas less dilute pop-1(RNAi) bacteria corresponded to a larger increase in seam cell number (e.g. 33.8 seam cell at a 50:50 dilution of lacZ(RNAi):pop-1(RNAi)) (Figure 1B). Since our goal was to obtain an intermediate seam cell phenocopy, we proceeded with our empty vector versus lacZ comparison using a 50:50 dilution ratio.
We next asked if an empty vector or a vector containing lacZ would affect seam cell output differently when diluted at the same ratio. Indeed, we saw that when pop-1(RNAi) bacteria were diluted 50% with lacZ, the worms resulted in significantly fewer seam cells (average, 33.8 seam cells) compared to dilution at the same ratio with empty vector (average, 49.4 seam cells) (Figure 1C). This result indicates that bacterially supplied lacZ dsRNA was more efficient in attenuating the pop-1 RNAi response compared to dsRNA produced from empty vector. Although lacZ dsRNA should not target the C. elegans transcriptome, we wanted to confirm that lacZ(RNAi) was not affecting pop-1 levels. To do this, we performed reverse-transcriptase PCR on worms subjected to RNAi of lacZ, pop-1, or empty pL4440 vector. As expected, we only saw a change in pop-1 expression in the pop-1(RNAi) condition (Figure 1D).
Together, our data show that bacterial mixing with a vector generating longer dsRNA serves to better dilute knockdown of a target gene compared to empty vector. The differences in effective dilution may be attributed to a similar number of dsRNA segments being produced from both the lacZ(RNAi) and pop-1(RNAi) vectors. In contrast, the minimal dsRNA produced by the empty vector(RNAi) may result in significantly less empty vector loaded RNAi riboproteins compared to target gene loaded riboproteins. In summary, the length of dsRNA generated by the diluting vector should be similar to the length of dsRNA produced by the target gene vector for effective bacterial mixing RNAi dilution.
Methods
Request a detailed protocolThe lacZ sequence was PCR amplified, inserted into pL4440, and sequence verified. RNAi constructs were expressed from E. coli HT115(DE3) bacteria, which were grown on plates containing 1 mM IPTG to induce T7 polymerase activity at convergent promoters (Timmons and Fire 1998). Simultaneous RNAi was carried out by growing the two different RNAi bacterial strains to the same optical density and mixing volumes in the ratios indicated before plating. RNAi was induced by L1 synchronized feeding (Timmons and Fire 1998) for 75 hours at 15°C and resulting L4 worms were immobilized using 1mM Levamisole on 5% agarose pads for live imaging. Six L4 worms not harvested for imaging were used to isolate RNA (Ly, Reid, and Snell 2015). Specifically, the six cleaned worms were transferred into 6ul of worm lysis buffer, frozen at -80°C then incubated in a thermocycler at 65 °C for 10 minute followed by 85 °C for 1 minute, to inactivate proteinase K. The worm lysate was then used immediately for semi-quantitative reverse-transcriptase PCR using SuperScript® III First-Strand Synthesis System with oligo(dT) followed by 25 rounds of pop-1 cDNA-specific amplification. In the genome, a large intron is located between primer sites, which distinguishes genomic DNA from cDNA.
Reagents
Strain JR667 [unc-119(e2498::Tc1) III; wIs51 (SCMp::GFP + unc-119) V]
SuperScript® III First-Strand Synthesis System
Primers
Primer 5’ to 3’ sequence
lacZ F: AACGTCGTGACTGGGAAAA
lacZ R: GACCTGACCATGCAGAGGAT
pop-1 mRNA F: ATGATGGCCGACGAAGAGCTCGGCG
pop-1 mRNA R: CATCGTATGCATCATTGCTGCTTG
Acknowledgments
We thank the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs [grant number P40 OD010440] and lab of Joel Rothman for providing strain JR667. We also thank the Kimble lab for generating the pop-1(RNAi) vector used.
References
Funding
This work was funded by the NIH grant NIGMS 1R01GM114007-01 and the NSF grant IOS-1917169
Reviewed By
Lisa TimmonsHistory
Received: January 30, 2020Revision received: May 21, 2020
Accepted: June 8, 2020
Published: July 1, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Bekas, KN; Phillips, BT (2020). Generating reliable hypomorphic phenocopies by RNAi using long dsRNA as diluent. microPublication Biology. 10.17912/micropub.biology.000269.Download: RIS BibTeX