BioSystems & Integrative Sciences Institute (BioISI), Plant Functional Biology Center, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal
Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, Rua Campo Alegre, 4169-007 Porto, Portugal
Description
Carbohydrate metabolism needs to be tightly regulated in order to ensure an organism’s survival. For this reason, several sensing mechanisms are in permanent alert to monitor and maintain homeostatic levels of sugars (Li and Sheen, 2016). For instance, glucose cellular levels can be sensed by different signaling proteins, including the Hexokinase 1 (HXK1), a moonlighting protein with both metabolic (glucose phosphorylation) and sensing/transduction functions (Moore et al., 2003). In a situation of excess glucose, HXK1 forms a nuclear complex that directly inhibits photosynthesis-related genes, reducing the synthesis of more glucose (Cho et al., 2006). Interestingly, HXK1 loss-of-function mutants lose the capacity to sense external sugar and, consequently, are capable of germinating in high percentages of external sugar supplementation. Indeed, the reference HXK1 mutant gin2-1, that harbors a nonsense mutation (Q432*) in Arabidopsis thaliana ecotype Landsberg erecta (Ler), was obtained from a forward screening for glucose insensitive (gin) mutants that do not suffer post-germinative growth arrest (PGGA) on a 6% glucose MS medium (Moore et al., 2003). Broadly, a myriad of phenotypic defects have been described for HXK1 mutants, highlighting the protein’s importance in the regulation of plant development (Aguilera-Alvarado and Sanchez-Nieto, 2017; Van Dingenen et al., 2019). Considering HXK1’s importance to plant fitness and development, one expects its activity to be tightly controlled.
During a previous study (Castro et al., 2015), we aimed to link carbohydrate signaling with the post-translational modification by the small peptide Small Ubiquitin-like Modifier (SUMO). SUMO conjugation is a reversible and rapid modification that can regulate the activity, localization and stability of target proteins (Castro et al., 2012). The covalent attachment of SUMO to a target (i.e. sumoylation), is mediated by an enzymatic cascade that sequentially involves DSP proteases (for SUMO maturation), E1 activating enzymes, E2 conjugating enzymes, and E3 ligases (Castro et al., 2012; Castro et al., 2018a). SIZ1 is an E3 ligase that is critical to enhance SUMO conjugation, especially in response to environmental stressors. Accordantly, SIZ1 loss-of-function mutants are not only developmentally compromised, but also hypersensitive to several environmental stress conditions (Castro et al., 2012).
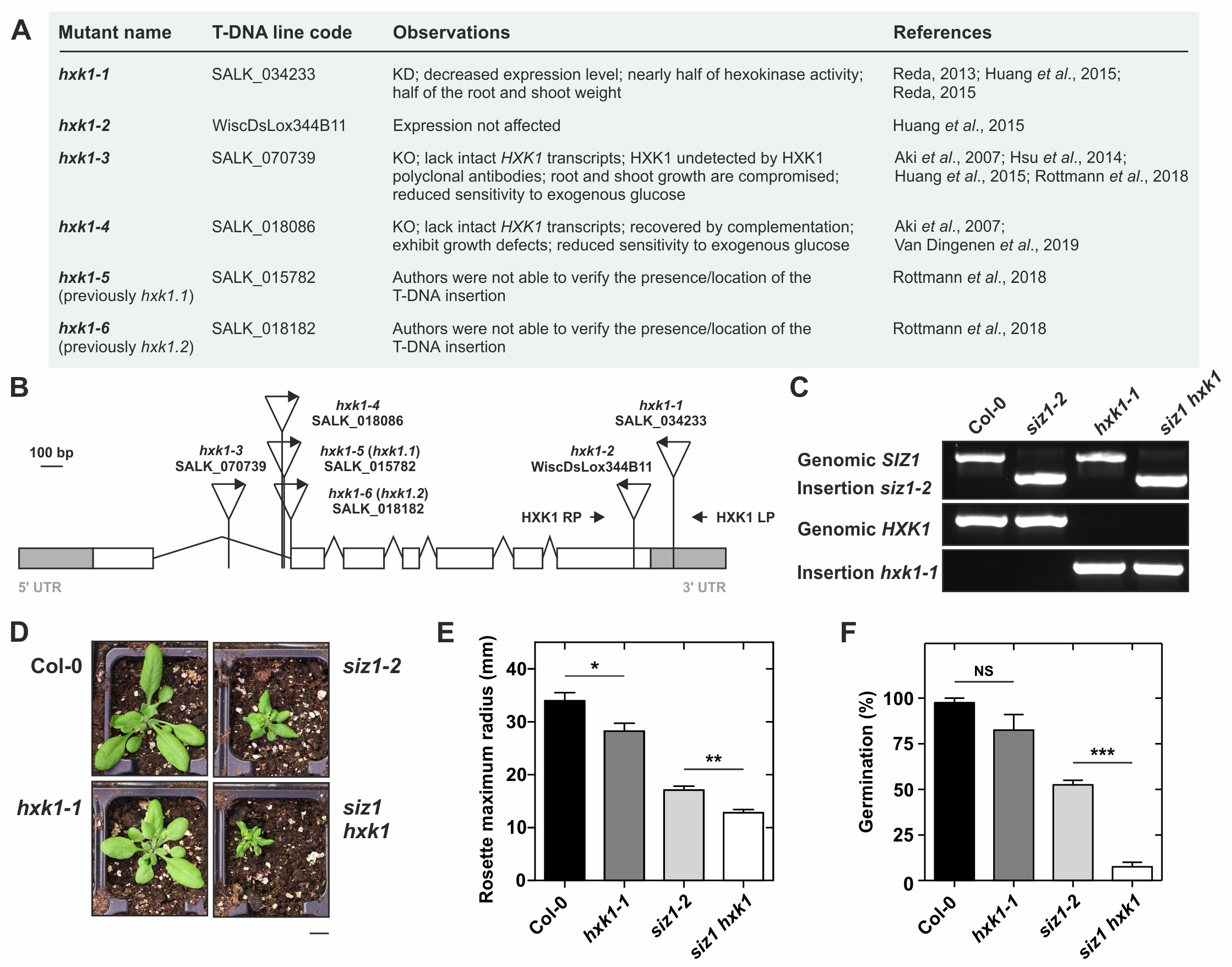
Previously, we reported that the siz1 mutation results in sugar hypersensitivity in its osmotic stress component, and also in the signaling effect of sugars leading up to PGGA (Castro et al., 2015; Castro et al., 2018b). Moreover, siz1 presents less glucose, fructose, sucrose and starch, which is likely correlated with enhanced expression of sugar degradation genes (Park et al., 2012; Tomanov et al., 2014; Castro et al., 2015). Interestingly, the glucose sensor transcript HXK1 and HXK-metabolic pathway marker genes were differentially expressed in siz1, and for this reason, we aimed to test for the presence of epistatic relationships between SIZ1 and HXK1. Consequently, we crossed siz1-2 with the previously used HXK1 T-DNA insertion mutant SALK_034233 (Reda, 2013; Huang et al., 2015; Reda, 2015), hereafter designated hxk1-1 (Fig. 1A,B). Since the siz1-2 mutant is a SALK line in the Columbia-0 (Col-0) background, we also opted for a SALK mutant for HXK1, instead of the well-characterized gin2-1 (in Ler background). During our inspection for Col-based insertion mutant lines present in the literature, we came across conflicting mutant allele nomenclatures. As such, in Fig. 1A,B we try to summarize existing knowledge, and suggest a novel nomenclature for several HXK1 insertion mutant lines for the community. In the F2 generation, hxk1-1 plants were genotyped using the primers displayed in Fig. 1B, resulting in the identification of the double mutant siz1-2 hxk1-1 (siz1 hxk1) (Fig. 1C). The hxk1-1 mutant was smaller than the wild-type, similar to other hxk1 alleles (Reda, 2013; Huang et al., 2015; Reda, 2015; Van Dingenen et al., 2019). Most significantly, the double mutant siz1 hxk1 showed greater morphological defects than siz1-2 (Fig. 1D), which suggests that hxk1-1 might produce an additive effect to the dwarfism displayed by the siz1-2 mutant (Fig. 1E). The germination also seems additively affected in the double mutant with only a few seeds germinating and producing cotyledons (Fig. 1F). In comparison with other hxk1 alleles, hxk1-1 presents a milder developmental defect (Aki et al., 2007; Huang et al., 2015; Van Dingenen et al., 2019). Since the hxk1-1 T-DNA insertion is in the 3’UTR (Fig. 1B) it is most likely a knockdown mutant allele. In support, Huang et al. (2015) showed a slight decrease of HXK1 expression in the hxk1-1 line. Albeit these results suggest that hxk1-1 is not a complete loss-of-function mutant, Reda (2013, 2015) stated that it lacks HXK1 expression (not shown by the author) and showed that this same mutant line is compromised in hexokinase activity by ~45%. This HXK1 knockdown mutant may be useful for genetic strategies where hxk mutants are introgressed into mutant backgrounds that are also severely pleiotropic, as is the current case of siz1. Nevertheless, the epistatic relationship between HXK1 and SIZ1 should be further clarified using null alleles of HXK1.
Collectively, these results support that SIZ1 and HXK1 are likely to be involved in independent growth control pathways. This also reinforces the idea that the HXK1-signaling pathway is unlikely to involve SIZ1 (Castro et al., 2015). However, the possible involvement of SUMO in the control of HXK1 activity, especially its nuclear localization and regulation of gene expression, cannot be discarded. This is supported by the presence of evolutionarily conserved sumoylation motifs in this protein family (Castro et al., 2020). Moreover, HXK1 was detected in a yeast-two-hybrid screening for SUMO E2 conjugase enzyme (SCE) interacting partners, and was sumoylated in a heterologous bacterial system (Elrouby and Coupland, 2010), which may indicate the existence of pathways alternative to SIZ1. This would mean either direct SUMO modification by E2 conjugases, or the action of alternative E3 ligases, in the modulation of HXK1 activities.
Methods
Request a detailed protocolPlant handling: Arabidopsis thaliana mutant lines are in the ecotype Columbia-0 background. The T-DNA insertion mutants siz1-2 (SALK_065397) and hxk1-1 (SALK_034233) were ordered from the NASC European Arabidopsis Stock Centre (arabidopsis.info) and confirmed by genotyping using the following primers: LBb1.3 5’-ATTTTGCCGATTTCGGAAC-3’; SIZ1-2 RP 5’-CACGACAGATGAAGCATTGTG-3’; SIZ1-2 LP 5’-GAGCTGAAGCATCTGGTTTTG-3’; HXK1 RP 5’-AGCTCGTCTCTCTGCTGCTGGA-3’; HXK1 LP 5’-CAGAACTCCAGTGAAGTGAGCTTTGA-3’. Synchronized Arabidopsis seeds were stratified, surface sterilized and sown onto MS medium as previously described by Castro et al. (2015). Plants were grown in growth chambers with a 16 h light/8 h dark cycle under cool white light (80 µE m-2s-1 light intensity) at 22-24ºC. For standard growth, in-vitro-grown 7-day-old seedlings were transferred to a soil to vermiculite (4:1) mixture and maintained with regular watering.
Phenotyping: Rosette size was measured using the ImageJ software (https://imagej.nih.gov/ij/). Germination percentages were determined 10 days after plating onto half-strength MS medium, by scoring green cotyledon appearance using a stereomicroscope.
References
Funding
Financial support was provided by Fundação para a Ciência e Tecnologia (FCT/MCTES) to H.A. [CEECIND/00399/2017/CP1423/CT0004] and from FCT/MCTES, Fundo Europeu de Desenvolvimento Regional (FEDER), and COMPETE – POCI – Programa Operacional Competividade e Internacionalização, to P.H.C. [PTDC/BAA-AGR/31122/2017, POCI-01-0145-FEDER- 031122].
Reviewed By
AnonymousHistory
Received: February 13, 2020Revision received: May 29, 2020
Accepted: June 16, 2020
Published: June 29, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Castro, PH; Verde, N; Azevedo, H (2020). Arabidopsis thaliana growth is independently controlled by the SUMO E3 ligase SIZ1 and Hexokinase 1. microPublication Biology. 10.17912/micropub.biology.000270.Download: RIS BibTeX