Cardiac Sciences, Libin Cardiology Institute, Cumming School of Medicine, University of Calgary, Calgary, Alberta CANADA
Description
The human LEMD2 protein and its homologs in other animals are associated with the inner nuclear membrane, the nuclear lamina and with functions such as chromatin organization and nuclear repair (Barton et al. 2015). The human mutation (c.38T>G; L13R) changes a single amino acid in the highly conserved LEM domain and, when homozygous, is associated with juvenile cataracts and with a greatly increased incidence of early onset cardiac arrest (Shokeir and Lowry 1985; Boone et al. 2016; Abdelfatah et al. 2019). The carrier frequency of this mutation in the North American Hutterite population is estimated to be as high as 12% (Abdelfatah et al. 2019).
The LEMD2 homolog in C. elegans is LEM-2. The lem-2 gene has been well characterized and appears to be largely redundant with the gene emr-1 (Lee et al. 2000; Liu et al. 2003; Barkan et al. 2012; Cohen-Fix and Askjaer 2017). Although lem-2 knockouts, whether by gene deletion or by administration of RNAi, show only mild phenotypes, ablation of both lem-2 and emr-1 genes causes complete lethality: if both zygotic and maternal contributions are removed, animals arrest as early embryos; maternally rescued animals arrest at ~ the L2 larval stage (Lee et al. 2000; Liu et al. 2003; Barkan et al. 2012; Cohen-Fix and Askjaer 2017). We have reconstructed the “Hutterite-type cataract/cardiomyopathy” mutation in the C. elegans lem-2 gene and now compare mutant phenotypes to the phenotypes produced by complete lem-2 knockouts. A longer term aim will be to exploit this reconstructed mutation in C. elegans to identify LEM-2 interacting factors, both biochemically and genetically.
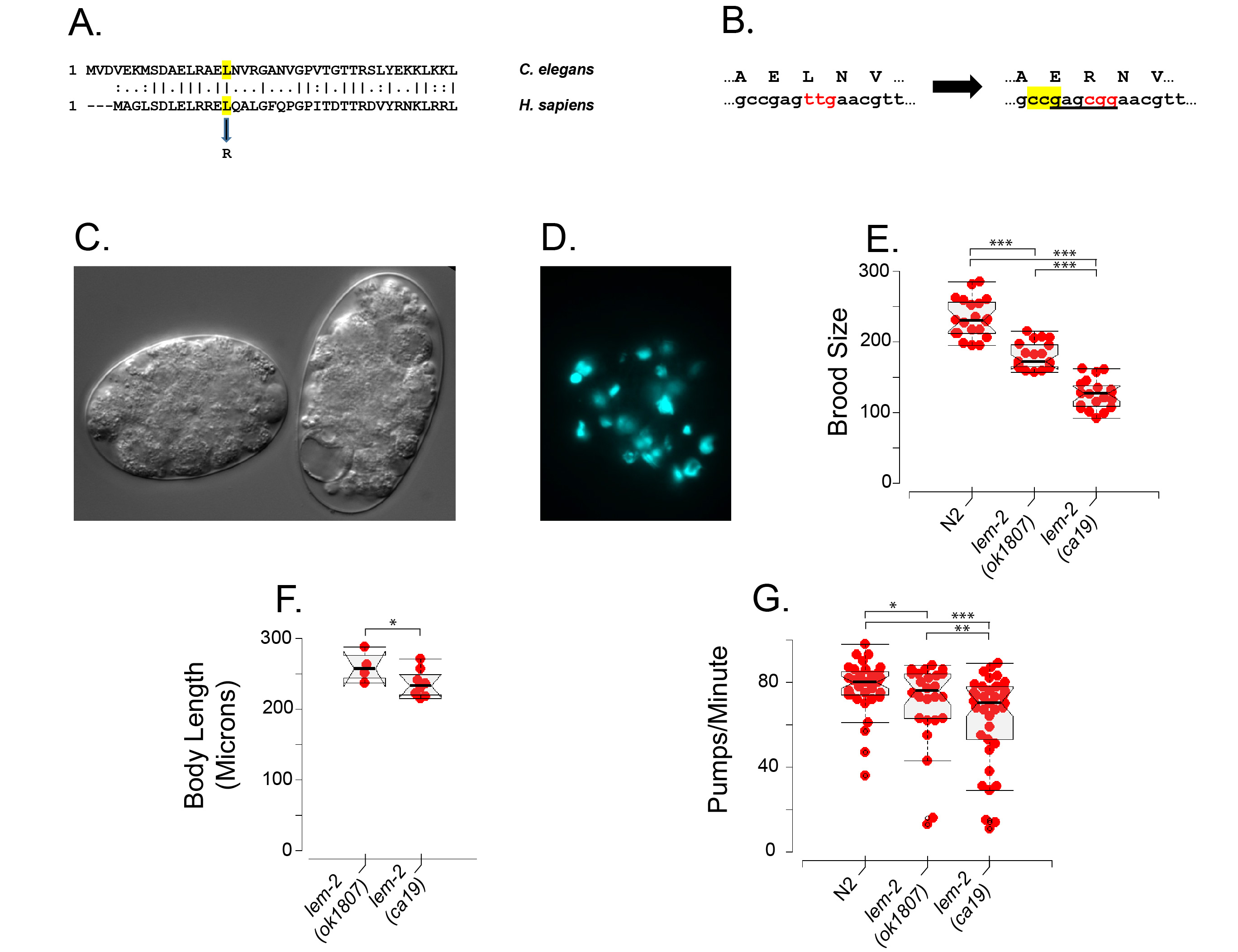
A partial sequence alignment (Figure 1A) shows that amino acid leucine 16 in the C. elegans LEM-2 protein corresponds to amino acid leucine 13 in human LEMD2. As shown in Figure 1B, we used CRISPR-Cas9 methods (Dokshin et al. 2018) to convert C. elegans leucine 16 to Arginine 16 (codon change from TTG to CGG); the mutation, designated lem-2(ca19), introduced a BsrbI restriction site that was used to follow the mutant gene through genetic crosses, including two initial outcrosses. The lem-2(ca19) mutation acts similarly to a complete lem-2 gene knockout: treating the otherwise quite healthy strain JM311 lem-2(ca19) with emr-1 RNAi by feeding (Kamath and Ahringer 2003; Kamath et al. 2003) leads to 100 % lethality, as previously reported for the lem-2 deletion allele tm1582 (Barkan et al. 2012) and that we now also confirm for the lem-2 deletion allele ok1807 used as our positive control. The key phenotype (Figure 1C) is embryonic arrest with fewer than 100 cells, frequent vacuoles and no obvious sign of differentiation (standard gut granule birefringence assay); a typical arrested embryo stained with DAPI (1 µg/ml) shows irregular condensed nuclei and occasional anaphase bridges (Figure 1D), much as has been previously reported for the complete lem-2(tm1582) knockout (Barkan et al. 2012) and as we now also confirm for the lem-2(ok1807) deletion. Administering emr-1 RNAi by injection (generally recognized as more effective than feeding) into JM311 quickly leads to maternal sterility, consistent with LEM-2/EMR-1 requirements in the germline. At this point, we conclude that the lem-2(ca19) mutation behaves as if it approximates a null.
We now looked more closely at the phenotypes of the lem-2 mutations in the presence of wildtype emr-1 function. Figure 1E shows that both lem-2 alleles ok1807 and ca19 show decreased brood size compared to N2 wildtype animals. However, the ca19 brood size is significantly lower than is the brood size of the complete lem-2 knockout, i.e., the ca19 phenotype is “worse” than the null phenotype. Thus ca19 could be described as a mild antimorph (Muller 1932) and this antimorph classification will be explained and defended below. A mild antimorphic nature of allele ca19 is also revealed by measurements of the pure zygotic effect of lem-2 loss. Following the scheme described in (Barkan et al. 2012), we produced a balanced strain in which lem-2(ca19) was homozygous and the emr-1(gk119) deletion was heterozygous, balanced with the chromosomal translocation hT2green (see Methods for full genotype). 1/16 of the offspring of this balanced strain are homozygous for both lem-2(ca19) and emr-1(gk119) and arrest as non-Green L2 larvae. As shown in Figure 1F, the nose-to-tail length of arrested emr-1(gk119); lem-1(ca19) larvae) produced by emr-1(+) mothers is detectably lower than for the control emr-1(gk110); lem-2(ok1807) complete knockout larvae, again consistent with lem-2(ca19) behaving as a mild antimorph. Figure 1G shows that both lem-2 mutations show a slightly lower pharyngeal pumping rate; once again, the ca19 phenotype is slightly more severe than that of the complete gene knockout. The slower pumping rates shown by both mutants agree with previous observations made with the lem-2(tm1582) allele (Barkan et al. 2012).
In summary, we have produced a mutation in the C. elegans lem-2 gene reconstructing a LEMD2 mutation that causes juvenile cataracts and premature cardiac arrest in the North American Hutterite population. Our main conclusion is that, in C. elegans, this single amino acid mutation acts similarly to a complete loss of function mutation; however, it also appears to show a mild antimorphic character. LEM-domain proteins are known to be multifunctional, binding to other proteins (e.g. BAF and lamins), the nuclear membrane and even DNA (Barton et al. 2015). Thus, if the ca19 mutation compromises one LEM-2 function but not others, the mutant protein could form non-productive complexes that could interfere with wildtype function or with the redundant function of EMR-1, in other words act as an antimorph; we have not tried to test this model by assessing phenotypes of heterozygotes. In any case, even if this mutation can act like an antimorph in C. elegans, there is no guarantee that the corresponding mutation acts as an antimorph in humans; for example, molecular interactions with other components of the inner nuclear membrane could be different in humans and in worms.
Methods
Request a detailed protocolStrain JM311 lem-2(ca19) was produced as described above (Figure 1B and (Dokshin et al. 2018)), including two outcrosses to N2 wildtype worms and validation by sequencing of PCR amplified fragments. Strain VC1317 lem-2(ok1807) was obtained from the Caenorhabditis Genetics Centre and outcrossed once (the strain designation was not changed). To assess the phenotype of maternally rescued emr-1; lem-2 larvae, we constructed strains in which the lem-2 allele was homozygous but the emr-1 deletion allele was heterozygous and balanced by a reciprocal translocation for which we use the shorthand hT2green. The proper designation of hT2green is hT2[bli-4(e937) let-?(q782) qIs48] (I;III) where the integrated chromosomal insertion qIs48 [Pmyo-2::gfp; Ppes-10::gfp; Pges-1::gfp] results in GFP expression. The relevant strains used in this experiment are as follows: JM312 emr-1(gk119)/hT2green I; lem-2(ok1807) II; +/hT2green III. JM313 emr-1(gk119)/hT2green I; lem-2(ca19) II; +/hT2green III. To measure the tip-to-tail length of arrested larvae, small non-Green animals (picked at a time when the rescued Green animals on the plate were young adults) were suspended in egg buffer containing 0.2% Tricaine + 0.02% Tetramisole + 5mM Sodium Azide and mounted on agarose pads; DIC images were analyzed using ImageJ. Pumping rates were measured at room temperature with one day adults in the presence of 10mM serotonin in 10% M9 buffer mixed with an equal volume of overnight culture of E. coli OP50 (Weeks et al. 2018). Pumping rates were measured by video recording at a magnification of 20X and analyzed at slower frame rates. RNAi by feeding was performed as described by (Kamath and Ahringer 2003), using library clone M01D7.6. Double stranded RNA was made by in vitro transcription of the same plasmid and injected at a concentration of 1 mg/ml as previously described (Fukushige et al. 2005; Goszczynski and McGhee 2005).
Reagents
Strains JM311, JM312 and JM313 will be made available at the Caenorhabditis Genetics Center.
References
Funding
The authors gratefully acknowledge funding support from the following organizations: The Canadian “Rare Diseases:Models and Mechanisms” Network (RDMM), the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Alberta Children’s Hospital Research Institute (ACHRI). Some strains were provided by the Caenorhabditis Genetics Centre (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
AnonymousHistory
Received: June 15, 2020Accepted: June 23, 2020
Published: June 29, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
AlKhaleefa, A; Snider, FL; Duff, HJ; McGhee, JD (2020). Using the C. elegans lem-2 Gene to Reconstruct the Human LEMD2 Mutation Associated with Hutterite-type Cataract/Cardiomyopathy. microPublication Biology. 10.17912/micropub.biology.000273.Download: RIS BibTeX