InVivo Biosystems (formerly NemaMetrix, Inc.), Eugene, OR 97402, USA
Division of Gerontology and Geriatric Medicine, Department of Medicine, University of Washington, Seattle, WA 98104, USA
Description
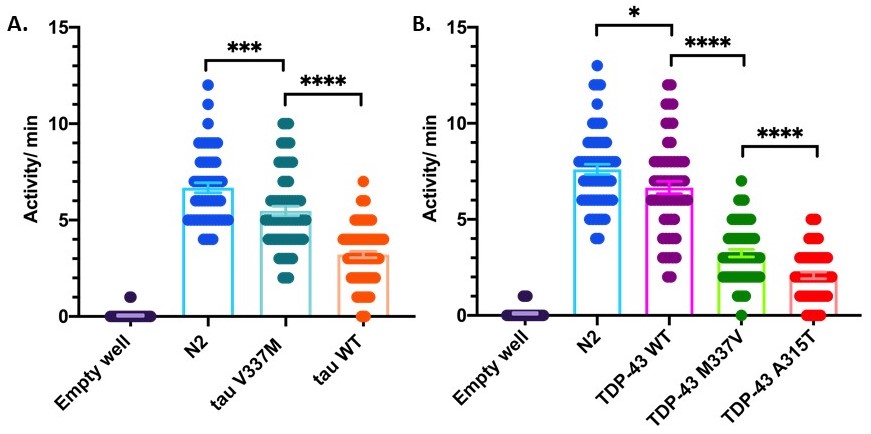
Aggregates of the protein tau are the hallmark of several neurodegenerative diseases including Alzheimer’s disease, frontotemporal lobar degeneration (FTLD-tau), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), Pick’s disease, and chronic traumatic encephalopathy (CTE) (VandeVrede, Boxer et al. 2020). Mutations in the gene coding for tau, MAPT, can cause FTLD-tau, directly linking tau dysfunction with disease (Dickson, Kouri et al. 2011). Another protein, TDP-43, comprises aggregates which are the primary hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD-TDP), and mutations in the gene coding for TDP-43, TARDBP, can cause disease (Kawakami, Arai et al. 2019). To model tau or TDP-43 proteinopathies, transgenic C. elegans have been generated that express the full-length human protein pan-neuronally. These worms exhibit significant uncoordinated movement on plates and impaired thrashing in liquid (Kraemer, Zhang et al. 2003; Liachko, Guthrie et al. 2010). However, tau- and TDP-43- expressing worms are not paralyzed; they still move their heads and have some motility on the plate (coiling, crawling with tail-drag, head swinging) which are not captured in standard crawling or thrashing assays. To assay differences in total activity, we used a WMicroTracker ARENA System (Phylumtech, AR and InVivo Biosystems, USA). The ARENA captures population level activity data by relying on optical interferometry, which uses a large array of infrared LED microbeams to detect both the movement and position of worms on a culture plate. Disruption of an LED microbeam by worm movement is recorded by repeat scans of the 6-well culture plate, and allows for real-time processing. The software identifies changes in the location of disrupted beams between scans and assigns an activity score based on differences identified between each consecutive scan (Simonetta SH). Both tau- and TDP-43- expressing worms had significantly less activity per minute than N2 (Figure 1). Further, we found the ARENA- assessed activity data recapitulated the relative severity of phenotypes among the strains as measured by motility assays. For example, both CK10 (tau V337M) and CK144 (tau WT) have significantly uncoordinated movement when crawling or thrashing in liquid, with CK144 having worse motility than CK10, due to its much higher burden of total tau protein expressed (Kraemer, Zhang et al. 2003). Likewise, CK410 (TDP-43 WT) worms have slightly impaired motility compared with N2 when crawling on a plate, CK423 (TDP-43 M337V) are severely uncoordinated, and CK426 (TDP-43 A315T) have the most severe uncoordinated phenotype. The relative toxicities of these strains stem from the effects of the mutations, as TDP-43 protein expression is relatively even among these transgenic strains (Liachko, Guthrie et al. 2010). Interestingly, the ARENA captures activity of these severely uncoordinated worms that move poorly in motility assays such as crawling on an NGM plate or thrashing in liquid (Kraemer, Zhang et al. 2003; Liachko, Guthrie et al. 2010). Therefore, ARENA assessment of aggregate activity may be a more accurate metric for capturing non-locomotor movement of C. elegans that are severely uncoordinated.
Methods
Request a detailed protocolARENA Tracking
All worms used were staged by way of timed egglay, and grown at 16oC on NGM plates seeded with OP50 E. coli to day 1 adult. The experiment was performed in a 6-well plate prepared with 3.5mL NGM per well and seeded with OP50. The worms were picked from stock plates into a prepared 6-well plate, 30 worms per strain, each strain in a unique well. Recordings were performed at room temperature (approximately 22oC). The plate was placed in the ARENA 15 minutes before starting the run to allow worms to acclimate. 14 activity scores were collected per strain over 60 minutes. Statistical analysis was performed with GraphPad Prism 8.
Reagents
Strains Used
N2 Bristol
CK10 bkIs10 [aex-3p::tau(V337M 4R1N); myo-2p::dsRED] III
CK144 bkIs144 [aex-3p::tau(WT 4R1N); myo-2p::dsRED] IV
CK410 bkIs410 [snb-1p::TDP-43(WT); myo-2p::dsRED] II
CK423 bkIs423 [snb-1p::TDP-43(M337V); myo-2p::dsRED] IV
CK426 bkIs426 [snb-1p::TDP-43(A315T); myo-2p::dsRED] II
Acknowledgments
We thank Dr. Brian Kraemer for strains and expert advice. We thank the Department of Veterans Affairs for funding.
References
Funding
Department of Veterans Affairs Merit Review Grant #I01BX004044 to N.L.
Reviewed By
Alex ParkerHistory
Received: June 22, 2020Revision received: July 13, 2020
Accepted: July 16, 2020
Published: July 16, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Currey, HN; Malinkevich, A; Melquist, P; Liachko, NF (2020). ARENA-based activity profiling of tau and TDP-43 transgenic C. elegans. microPublication Biology. 10.17912/micropub.biology.000278.Download: RIS BibTeX