Description
In Drosophila melanogaster, approximately 150 so-called clock neurons express the molecular components of the circadian clock mechanism. The activity of these neurons enables the fly to anticipate and track cyclic changes in their environment and to respond to them accordingly. The core of the circadian clock is built by two interlocked transcriptional feedback loops. In the first loop, the transcription factors CLOCK (CLK) and CYCLE (CYC) initiate the expression of the core clock proteins PERIOD (PER) and TIMELESS (TIM), which are in turn repressing the activity of CLK and CYC as transcriptional activators, resulting in a cyclic accumulation of PER and TIM (reviewed in Hermann-Luibl and Helfrich-Förster, 2015). In light:dark cycles, the intracellular photoreceptor Cryptochrome (CRY) mediates light information to the core clock and resets the molecular mechanism, thereby synchronizing the clock every day at sunrise to the 24h oscillation (reviewed by Helfrich-Förster, 2020). However, in constant darkness, the oscillation of the clock proteins PER and TIM cycles with the endogenous period, which is close to, but not exactly 24hrs.
A commonly used read-out for the molecular oscillations are immunohistochemical assays to quantify the clock protein abundance over time. For this endeavor, the researcher has to dissect and analyze a large number of animals, which is very labor-intensive and time-consuming. A standard timeseries in Drosophila chronobiology takes at least 80 flies per genotype, when the sampling occurs every 3hrs (8 timepoints, 10 flies per timepoint).
In 2011, the lab of Herman Wijnen was the first to describe a procedure which enables to assess the clock protein cycling of single clock neurons in individual brains of Drosophila by imaging PER-Luciferase (PER-LUC) activity (Sellix et al., 2010). Even though this technique has the potential to overcome the labour-intensive timeseries sampling for immunohistochemical experiments, only one more study utilized the single-cell luciferase imaging approach resulting in two publications (Roberts et al., 2015 and Roberts et al., 2016). We believe that this is on one hand partially caused by the complicated cultivation protocols available and on the other hand that the two mentioned studies have been performed on custom-built microscope setups, which might deter potential users. (Author comment: An additional study using a similar experimental setup as described in this article has been uploaded to the bioRxiv preprint server while this article was under review (Versteven et al., 2020)).
In this article, we briefly describe a culturing protocol for explanted Drosophila brains, which is sufficient to keep the brains alive and healthy for more than a week, even without partial substitution of the culture medium on a regular basis. We further used a commercial Olympus Luminoview (LV200) luminescence microscope equipped with an EMCCD camera (ImagEm X2 9100-23B, Hamamatsu Photonics) to monitor the PER-LUC expression of single clock neurons over several circadian cycles to show the viability of the explanted brains in an independent assay.
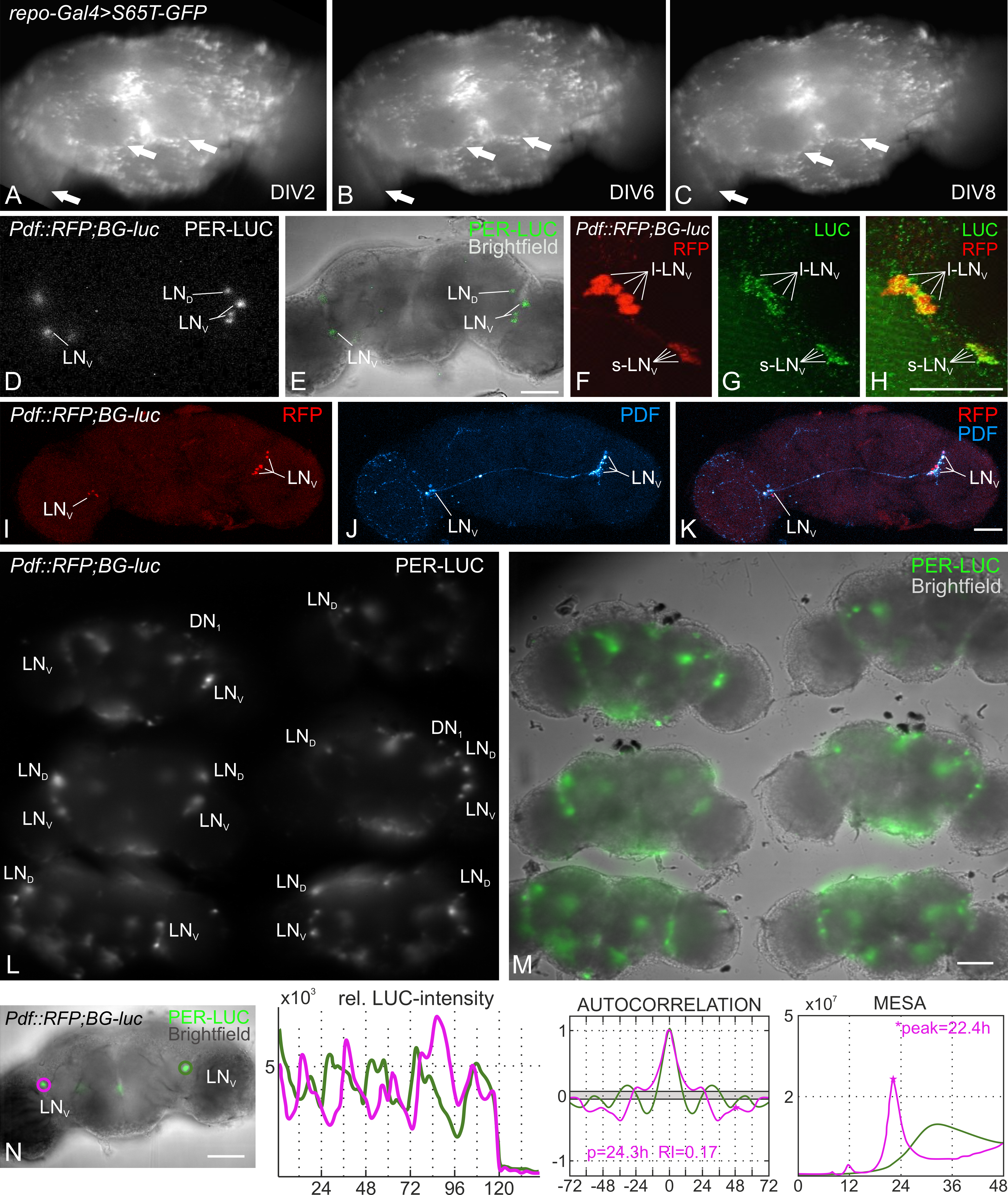
The flies for this experiment were reared under 12:12 hr light:dark-cycles on standard corn agar medium at 25°C with 60% relative air humidity. After dissection in ice cold Ca2+ free Ringer’s solution (see reagents), we placed the brains in a Poly-L-Lysine coated glass bottom petri dish (Greiner Bio-One) and added the culture medium. The culture medium contains 20% heat-inactivated fetal bovine serum, 1% Penicillin-Streptomycin and 0.75 µM Luciferin. Subsequently, the dish with the brain explants was transferred either to a culture incubator or to the luminescence microscope for imaging. For avoiding movement artifacts during long-term recording, we decided to culture the brain explants without partially substituting the culturing medium. To test the viability of these explants, we assessed pan-glial EGFP expression over the course of several days, since glia are essential for neuronal survival and suppression of cell death (Buchanan and Benzer, 1993; Xiong and Montell, 1995; Volkenhoff et al., 2015). We could not observe any dramatic loss of glial cells after eight days in vitro (DIV8) without adding fresh medium (Fig. 1 A-C). The majority of the targeted cells express EGFP on DIV8, and we therefore concluded that the explants are viable for at least eight days in culture. In the next step we assayed brains from the y w; Pdf::RFP1; BG-luc line (expressing PER-LUC in all clock neurons and RFP under the control of the Pdf promotor) under the LV200 luminescence microscope. The resolution of the setup is sufficient to identify single clock neurons by their PER-LUC expression by using either 30-fold or 60-fold super apochromat objectives and 300 s exposure time (UPLSAPO 30x, UPLSAPO 60x, Olympus life science; Fig. 1 D, and 1 L, respectively). The specificity of the y w; Pdf::RFP1; BG-luc line was confirmed by analyzing the native Pdf::mRFP1 expression and a separate staining against the firefly luciferase, which was colocalized in the Pigment Dispersing Factor expressing (PDF+) ventrolateral clock neurons (Fig. 1 F-H). We could further show that the staining for the circadian output factor PDF also worked after the brain had been imaged and kept in culture for eight consecutive days, showing that the tissue is indeed viable throughout the complete recording time (Fig. 1 I-K). Even though it was not possible to record z-stacks of the cultivated brains with the LV200 due to the long exposure time (300 s for one plane), the simultaneous recording of up to six synchronized brains is an option to increase the probability of recording several circadian clock clusters of different focal planes and/ or to increase sample size (Fig. 1 L, M).
The analysis of the recorded large ventrolateral clock neurons’ (l-LNv) PER-LUC rhythms via a MATLAB toolbox (Levine et al., 2002) identified a protein cycling with a circadian period of approximately 24hrs in one of the neurons (magenta circled neuron, Fig. 1 N). The period length estimated by MESA is shorter than the one obtained with autocorrelation analysis (22.4hrs and 24.3hrs, respectively; Fig. 1 N). In the other neuron both methods failed to detect rhythmicity, even though the oscillations appeared rhythmic (green circled neuron, Fig. 1 N). We estimated the period length by manually measuring the peak-to-peak intervals. Although the underlying rhythm is not significant, the manually calculated period length of approximately 23.6hrs falls into the circadian range. It is reported that the l-LNv would not cycle in constant darkness in the intact fly (Grima et al., 2004), but the aforementioned studies observe a PER cycling in those cells as well (Sellix et al., 2010, Roberts et al., 2015, Roberts et al., 2016). This is most likely due to the loss of inhibitory signals from the periphery (e.g. photoreceptors, Schlichting et al., 2016).
We therefore conclude that, depending on the study aim, the recording of luciferase reporter expression of explanted brains is a well-suited alternative to the classical work-intensive immunohistochemical time-course experiments and combines a higher resolution of the clock protein cycling together with a smaller number of animals that need to be studied.
Reagents
Fly lines:
y w; Pdf::RFP1; BG-luc (y w; P{Pdf-RFP.R}; P{BG-luc}); expresses mRFP1 under the control of the Pdf promotor and a PERIOD-LUCIFERASE fusion protein in all clock neurons. See Ruben et al. (2012) for information on the Pdf::mRFP1 construct and Stanewsky et al. (1997) for information on the BG-luc line.
Repo-Gal4 (w1118; P{w+m*=GAL4}repo/TM3,Sb1; Bloomington stock #7415); expresses GAL4 in glia cells. See Sepp et al. (2001) for further information.
UAS-S65T-GFP (w*; P{w+mC=UAS-GFP.S65T}egT10; Bloomington stock #1522); expresses GFP.S65T under the control of UAS.
Primary antibodies:
Anti-LUC (Thermo Fisher Scientific, RRID: AB_1076533); raised in mouse, binds to the Luciferase (LUC) enzyme. Used in a final concentration of 1:2000.
Anti-mCherry (Thermo Fisher Scientific, RRID: AB_2536611); raised in rat, binds to red fluorescent protein (RFP) and its derivates. Used in a final concentration of 1:2000.
Anti-PDF-C7 (DSHB, deposited by J. Blau in 2005, RRID: AB_760350); raised in mouse, binds to the pigment dispersing factor (PDF) neuropeptide expressed by the lateral clock neurons. Used in a final concentration of 1:4000.
Secondary antibodies:
AlexaFluor488 anti mouse (Thermo Fisher Scientific, RRID: AB_2534069); used in a final concentration of 1:200.
AlexaFluor568 anti rat (Thermo Fisher Scientific, RRID: AB_2534121); used in a final concentration of 1:200.
AlexaFluor647 anti mouse (Thermo Fisher Scientific, RRID: AB_2535804); used in a final concentration of 1:200.
Buffers and solutions:
Ca2+ free Ringer’s solution (70 mM NaCl, 5 mM KCl, 20 mM MgCl2, 10 mM NaHCO3, 120mM Sucrose, 5mM HEPES; recipe shared by André Klarsfeld and Serge Birman)
Culture medium (SM with 20% FBS, 1% Pen/Strep and 0.75µM Luciferin; recipe shared by André Klarsfeld and Serge Birman)
Fetal bovine serum (FBS, PAA #A15-101)
Luciferin (Biosynth #L-8220)
Paraformaldehyde 4% (PFA, Affymetrix #199431LT)
Penicillin/ Streptomycin (Pen/Strep, PAA #P11-010)
Phosphate buffered Saline (PBS, Sigma-Aldrich #P5493)
Poly-L-lysine (Sigma-Aldrich #P4707)
Schneider’s insect medium (SM, Sigma-Aldrich #S0146)
Glass bottom culture dish Greiner Bio-One #627861
Acknowledgments
We thank Prof. Dr. Ralf Stanewsky for kindly providing the yw; Pdf::mRFP1; BG-luc fly line and for the fruitful discussions on this project. We further thank Dr. Saskia Eck for pushing the establishment of the brain cultures and Thomas Schubert for language corrections.
References
Funding
The study was funded by the German Research Foundation (DFG), collaborative research center SFB1047 “Insect Timing”, project A3 (project #208233609).
Reviewed By
Taishi Yoshii and Steven MarygoldHistory
Received: June 15, 2020Revision received: July 10, 2020
Accepted: July 16, 2020
Published: July 22, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Schubert, FK; Helfrich-Förster, C; Rieger, D (2020). Single-cell resolution long-term luciferase imaging in cultivated Drosophila brains. microPublication Biology. 10.17912/micropub.biology.000280.Download: RIS BibTeX