MARCU-STAR Program
University of California, Riverside
Department of Molecular, Cell and Systems Biology
Description
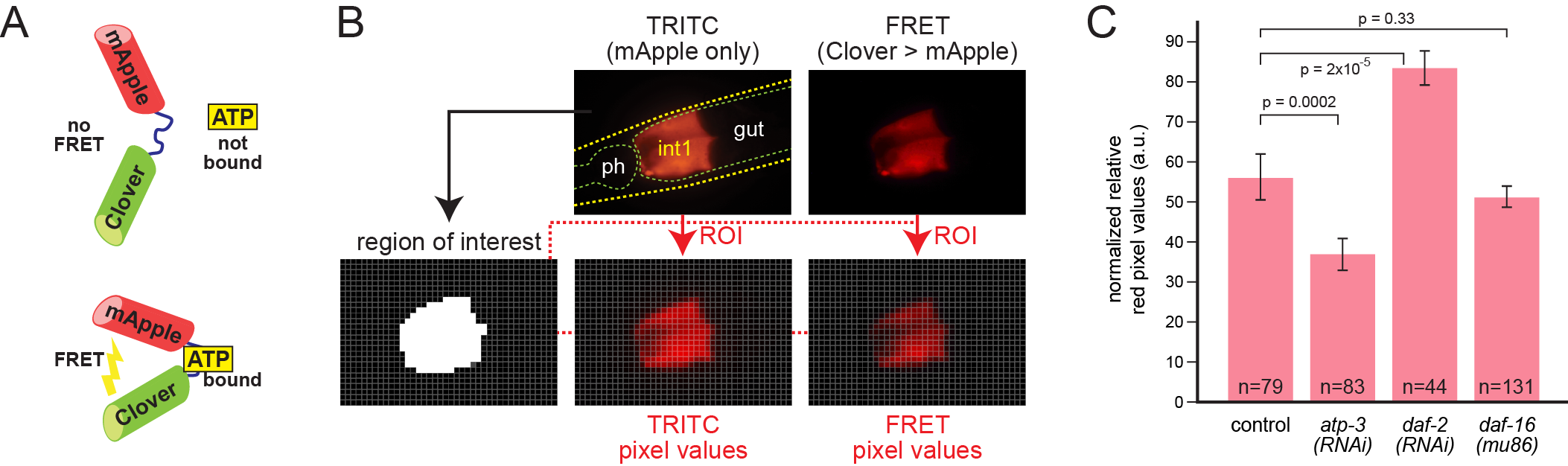
The C. elegans intestine is a major site of the generation and storage of chemical energy. Adenosine triphosphate (ATP) is a major molecule involved in energy transfer, hence regulation of intracellular ATP levels is a critical aspect of metabolism. Recently, ATP sensor proteins have been used to indirectly measure ATP levels in living cells (Zhang et al., 2018). These sensors are derived by inserting the ε subunit of the bacterial F0F1-ATP synthase between two fluorescent protein domains, allowing detection of protein bound to ATP by Förster Resonance Energy Transfer, or FRET (Imamura et al., 2009). Others have recently demonstrated use of FRET-based ATP sensors in the C. elegans pharynx and muscle (Tsuyama et al., 2013; Wang et al., 2019). Here we test the intestinal expression of a similar FRET-based ATP sensor derived from the green/red fluorochrome pair Clover/mApple (Figure 1A) (Mendelsohn et al., 2018). When the protein is bound to ATP, the two fluorochromes are brought into proximity, permitting emission from Clover to excite mApple (Imamura et al., 2009; Mendelsohn et al., 2018).
We made transgenic animals expressing the sensor under the control of the pept-1 promoter, which in arrays drove strongest expression in the anterior intestinal ring, int1 (Mendelsohn et al., 2018; Nehrke, 2003). We measured FRET by comparing red signal in image pairs as described in Figure 1B. Individual adult animals were imaged twice in the same focal plane. In the first image, mApple was detected by a TRITC filter set to reveal the sensor location independent of ATP levels. A second image was obtained using a filter set to excite Clover and detect red FRET emission from mApple. We devised an automated image analysis pipeline in Python to rapidly compute average ratios of FRET:mApple signal for each image pair and combine the data across multiple animals.
We tested three genetic conditions to evaluate whether the sensor produces similar relative ATP levels as previously determined by others (Fig. 1C). Knockdown of the nuclear-encoded ATP synthase subunit gene atp-3 by RNAi results in a decrease of 60-80%, and knockdown of daf-2, an increase of 100-200% in biochemical ATP levels in whole animals, depending on their age (Dillin et al., 2002). In contrast, mutation of daf-16 produces little or no change in ATP levels (Braeckman et al., 1999). Mutation of daf-2 produced a modest increase in muscle ATP levels after several days, measured by a similar FRET-based ATP sensor, ATeam (Wang et al., 2019). As shown in Fig. 1C, our results were qualitatively similar. However, the differences seen with the sensor, while statistically significant, are of smaller magnitude, i.e. only a 40% decrease in atp-3(RNAi) and a 50% increase in daf-2(RNAi). Conservatively, therefore, an in vivo ATP biosensor is best used to compare relative free ATP levels across genotypes and conditions, and not to directly infer actual biochemical changes.
Our results suggest that even with a modest low-cost fluorescence setup, an in vivo ATP FRET reporter can be a useful way to measure aspects of metabolic state in the C. elegans intestine. Improvements in expression of the reporter and the use of confocal microscopy are likely to further increase the usefulness of this system.
Methods
Request a detailed protocolConstruction of the sensor transgene. We obtained plasmids encoding the normal and kinase-dead versions of the novel Clover-ATP-mApple fusion protein (Mendelsohn et al., 2018). These were used for PCR and Gibson assembly to fuse 1407 bp of the pept-1 promoter to the sensor coding region and the 3’UTR of unc-54 from vector pPD95.67.
Strains and worm handling. unc-119(ed4) mutants were made transgenic for the pept-1-driven sensor and an unc-119(+) rescue plasmid by microinjection. Arrays were integrated to generate MS2495 strain carrying irIs158 (normal sensor) and MS2499 carrying irIs162 (kinase-dead sensor). RNAi was performed by feeding of the HT115 bacteria carrying the empty vector, daf-2 or atp-3 sequences using standard methods (Kamath and Ahringer, 2003).
Fluorescence microscopy. The anterior intestines of 1- to 2-day old adults were imaged on an Olympus BX-51 upright epifluorescence microscope equipped with a 100W mercury arc lamp and Canon EOS 77D camera with LMScope C-mount adapter. The mApple was detected using a Chroma 31002 TRITC filter set. For FRET detection we combined the HQ500/20x exciter and Q515lp beam splitter from a Chroma 41029 YFP filter set with a 635/20 emission bandpass filter (a gift from Dr. David Carter, UCR Microscopy Core). To minimize variability in fluorescence intensity, images for a set of experiments were acquired within a short time using the same mercury bulb and settings.
Image analysis. Images of size 2656×3984 pixels were taken at ISO 100 and ¼ sec exposure and saved in JPG format using Canon Digital Photo Professional. In our first studies, we opened image pairs as layers in Adobe Photoshop and use the control TRITC image to identify a region of interest. We would then extract pixel values of this region from both images. We developed a Python script to process image pairs automatically (available at https://github.com/MaduroMF/FRETcalc/releases/tag/1.0). The script subdivides the first image into 100×100 pixel blocks, and if the average red pixel value of a block is higher than a background of 50, average red pixel values are recorded from the same block between the control and FRET images. For each image pair the program computes both the average of the ratios obtained across the regions of interest, as well as the ratio of the average pixel values. For the histogram shown in the figure the latter was used to compute the means, standard error, and p values using a two-sided t test. The pixel ratio (r) values were scaled for the histogram by computing (r-0.04)/0.04*100. The subtracted ratio of 0.04 was obtained from measurements of background FRET from a kinase-dead version of the sensor.
Acknowledgments
We gratefully acknowledge Gladstone Institutes and Ken Nakamura, M.D., Ph.D for providing the plasmids used in this study, and acknowledge the contribution of Hiromi Imamura, Ph.D., Professor, Kyoto University, for the generation of the original ATP biosensor (Imamura et al., 2009).
References
Funding
This project was supported by a MARC U STAR grant to J.S., Award Number T34GM062756 from the National Institutes of Health.
Reviewed By
AnonymousHistory
Received: June 29, 2020Revision received: July 14, 2020
Accepted: July 22, 2020
Published: July 30, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Soto, J; Rivera, M; Broitman-Maduro, G; Maduro, MF (2020). Expression of a FRET-based ATP Biosensor in the C. elegans Intestine. microPublication Biology. 10.17912/micropub.biology.000284.Download: RIS BibTeX