Description
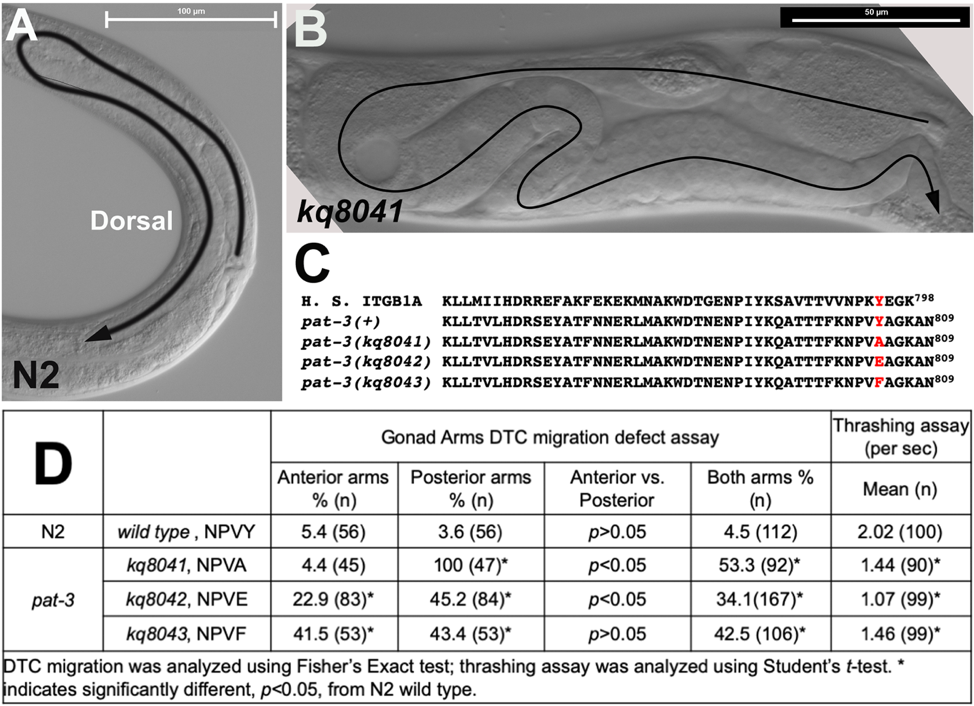
Integrin is a heterodimeric cell surface receptor for extracellular matrix proteins. C. elegans has two α integrin and one β integrin subunit. The β integrin PAT-3 contains two NPxY phospho-tyrosine motifs in the cytoplasmic domain (Figure 1C). The NPxY motif is known for interacting with talin and kindlins and plays essential roles in the bidirectional signaling of integrins (Hynes 2002). To investigate the role of tyrosine phosphorylation in the NPxY motifs, we mutated the tyrosine to different amino acids to mimic the physiological modifications. In this study, the membrane-distal NPxY was studied using genome editing with the CRISPR-Cas9 ribonucleoprotein complex system (Dickinson and Goldstein 2016). The membrane distal 801NPVY804 was engineered to three different forms, such as NPVF804 (phenylalanine), NPVA804 (alanine), or NPVE804 (glutamate). The NPVF804 is to mimic the non-phosphorylatable tyrosine (Xu et al. 2010). NPVA804 is to abolish the tyrosine residue (Chen et al. 2006). NPVE804 is to mimic the phosphorylation (Qiu et al. 2019), with the expectation that three CRISPR engineered lines would display defective motility and abnormal cell migration. None of the lines, however, showed lethality or noticeable abnormal appearance, but they showed defective gonad migration (Figures 1B and 1D) and mild movement defects (Figure 1D). All alleles displayed a significant percentage of DTC migration defects (>30%) (Figure 1D). It should be noted that the DTC Mig was observed more frequently in the posterior gonad in kq8041 (NPVA804) and kq8042 (NPVE804), while the DTC Mig of kq8043 (NPVF804) was equally detected in both gonad arms. All alleles showed the decrease in motility; the kq8042 was severer than other alleles. We believe our results are useful for in vivo analysis of integrin functions and cell-matrix interactions.
Methods
Request a detailed protocolThe CRISPR-Cas9 was used to edit the pat-3 locus to create the kq8041, kq8042, and kq8043 mutations. The potential crRNA sequence was present in exon 8 of the pat-3 gene covering the membrane-distal NPVY (Figure 1C). The DNA repair template spans 48 bases upstream and 38 bases downstream of the target site, tyrosine (Y804). Synonymous mutations modified many positions of codons in the crRNA target to identify the mutation. The repair DNA templates, crRNA, tracrRNA, and Cas9 nuclease were custom made from IDT Inc., Coralville, IA. A mixture of template DNA (PAT3Y2A, PAT3Y2E, or PAT3Y2F), crRNA (ZQPAT3B), tracrRNA (cat. no. 1073190), and Cas9 protein (cat. no. 1081058) was annealed at room temperature. The mixture was micro-injected into the syncytial gonad of N2 animals (P0) with dpy-10 co-CRISPR (Paix et al. 2015; Dickinson and Goldstein 2016). F1 animals with the Dpy phenotype were isolated, which displayed the mutation in single worm PCR genotyping with mutant specific primers (PCR-R-Y2F, PCR-R-Y2E, and PCR-R-Y2A) (Jansen et al. 1997). The Non-Dpy F2 homozygote was isolated; the animal displayed the mutation-specific PCR result but showed the absence of wild-type PCR result (the wild-type specific primer, PCR-WT-R-Y2). Homozygous mutants were crossed back to N2 two times. Three CRISPR edited mutant alleles, kq8041 (NPVA), kq8042 (NPVE), and kq8043 (NPVF), were generated. Each edited line underwent phenotype analyses. Briefly, mutant animals showed DTC migration defects under a Nomarski microscopy (Lee and Cram 2009). Morphology of U-shaped gonad arms was observed in L4 or young adult stage hermaphrodites. For thrashing assays, animals were placed in 10 μl M9 drops. The number of body bending in aqueous solution was measured for 10 seconds. A Fisher’s Exact test (DTC migration) and Student t-test (motility assay) was performed to confirm the statistical significance of assay results. Nucleotide sequences of repair template, PCR primers, and crRNA in this study are listed below.
| Repair template | Sequence (5’-3’) |
| Repair Y804F | GAGAACCCAATCTACAAACAGGCCACGACAACATTCAAGAACCCGGTTTTTGCAGGAAAAGCCAACTAAatagtttttatccttatatt |
| Repair Y804E | GAGAACCCAATCTACAAACAGGCCACGACAACATTCAAGAACCCGGTTGAAGCAGGAAAAGCCAACTAAatagtttttatccttatatt |
| Repair Y804A | GAGAACCCAATCTACAAACAGGCCACGACAACATTCAAGAACCCGGTTGCTGCAGGAAAAGCCAACTAAatagtttttatccttatatt |
Differentiated amino acids are italicized
| PCR primers | Sequence (5’-3’) | Used for |
| PCR-WT-R-Y2 | CCAGCGTATACTGGATTTTTA | wildtype specific |
| PCR-R-Y2F | GCAAAAACCGGGTTCTTG | Y804F specific |
| PCR-R-Y2E | CTTCAACCGGGTTCTTG | Y804E specific |
| PAT3MCRF | CATGATAGATCCGAATACGC | sequencing Forward |
| PCR-R-Y2A | CAGCAACCGGGTTCTTG | Y804A specific |
| PAT3R3UTR | acaatttatcgctaaatactcgtt | sequencing Reverse |
crRNA sequence
| ZQPAT3B | 5’-TTTAAAAATCCAGTATACGC-3’ | TGG (PAM target) |
Reagents
BU8041 pat-3(kq8041), BU8042 pat-3(kq8042), and BU8043 pat-3(kq8043) are available upon request.
Acknowledgments
These mutant lines were created during the course of BIO 4108 Cell and Developmental Biology Lab at Baylor University.
References
Funding
Funding for BIO 4108 was provided by Baylor University.
Reviewed By
AnonymousHistory
Received: June 25, 2020Revision received: July 30, 2020
Accepted: August 9, 2020
Published: August 15, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Hanna, J; Ramani, S; Williams, T; Anaya, R; Campion, N; Lopez, E; Williams, R; McIntire, J; Tran, N; Reyna, V; Zeng, J; Miller, S; Pancar, A; Qiu, Z; Lee, M (2020). Three alleles in the pat-3 locus of Caenorhabditis elegans: mutations in the membrane-distal NPxY phosphotyrosine motif. microPublication Biology. 10.17912/micropub.biology.000291.Download: RIS BibTeX