Description
Fluorescent markers are useful for identifying transgenic C. elegans after injection. In some cases, fluorophores are used to identify transgenics and to propagate animals with extra-chromosomal or integrated arrays (e.g., sur-5::gfp) (Gu et al. 1998). In other cases, fluorescent markers are used as visual markers to identify and later select against array animals. Such negative selection is used generate single-copy transgene insertions by plasmid injection, e.g., Mos1-mediated single-copy insertion (MosSCI)(Frøkjær-Jensen et al. 2008) or CRISPR/Cas9 (Dickinson et al. 2013). These methods generate targeted double-strand breaks, and transgenes are inserted by homologous recombination into specific locations. Selection schemes that rely on positive (e.g., cbr-unc-119, NeoR, HygroR)(Maduro and Pilgrim 1995; Giordano-Santini et al. 2010; Radman et al. 2013) and negative (e.g., fluorophores or the peel-1 toxin) (Frøkjær-Jensen et al. 2012) selection markers are frequently used to identify single-copy insertions. Negative peel-1 selection is under control of a heat-shock promoter (Phsp-16.41); after heat-shock, animals with arrays or dual insertions with the peel-1 transgene rapidly die (Seidel et al. 2011). peel-1 is convenient, but this selection has two significant drawbacks: the transgene is toxic in the absence of heat shock, resulting in fewer F1 progeny (Frøkjær-Jensen et al. 2012), and the induced lethality is often not fully penetrant resulting in false positives. Regardless of whether the peel-1 selection is used, we have advocated for the inclusion of all three red fluorescent markers expressed in neurons (Prab-3), body-wall muscle (Pmyo-3), and pharynx (Pmyo-2) because a single or even two markers were inefficient at avoiding false positives. These three selection markers are widely used and have been requested more than 300 times each from Addgene. However, adding three fluorescent markers to every injection mix is increasingly inconvenient as the mixes also contain plasmids encoding enzymes (Mos1 transposase or Cas9), sgRNAs, repair templates, and sometimes additional markers. Pharyngeal expression of the Pmyo-2 fluorophore is the easiest to identify on a fluorescence dissection microscope due to early embryonic expression and brightness, but the transgene is frequently toxic and is therefore not commonly injected at high concentrations. For these reasons, we sought to identify an improved co-injection marker. An ideal marker would be bright, non-toxic, expressed in a clearly identifiable tissue, and only require a single marker in the injection mix. Here we demonstrate that fluorescent transgenes containing the pan-muscular promoter from myosin light chain 1 (mlc-1) fulfill these criteria. The mlc-1 and the related mlc-2 promoter may also be generally useful to identify all (body wall, pharynx, vulval, stomato-intestinal, and anal depressor) muscles or for robust expression of transgenes (e.g., optogenetic sensors).
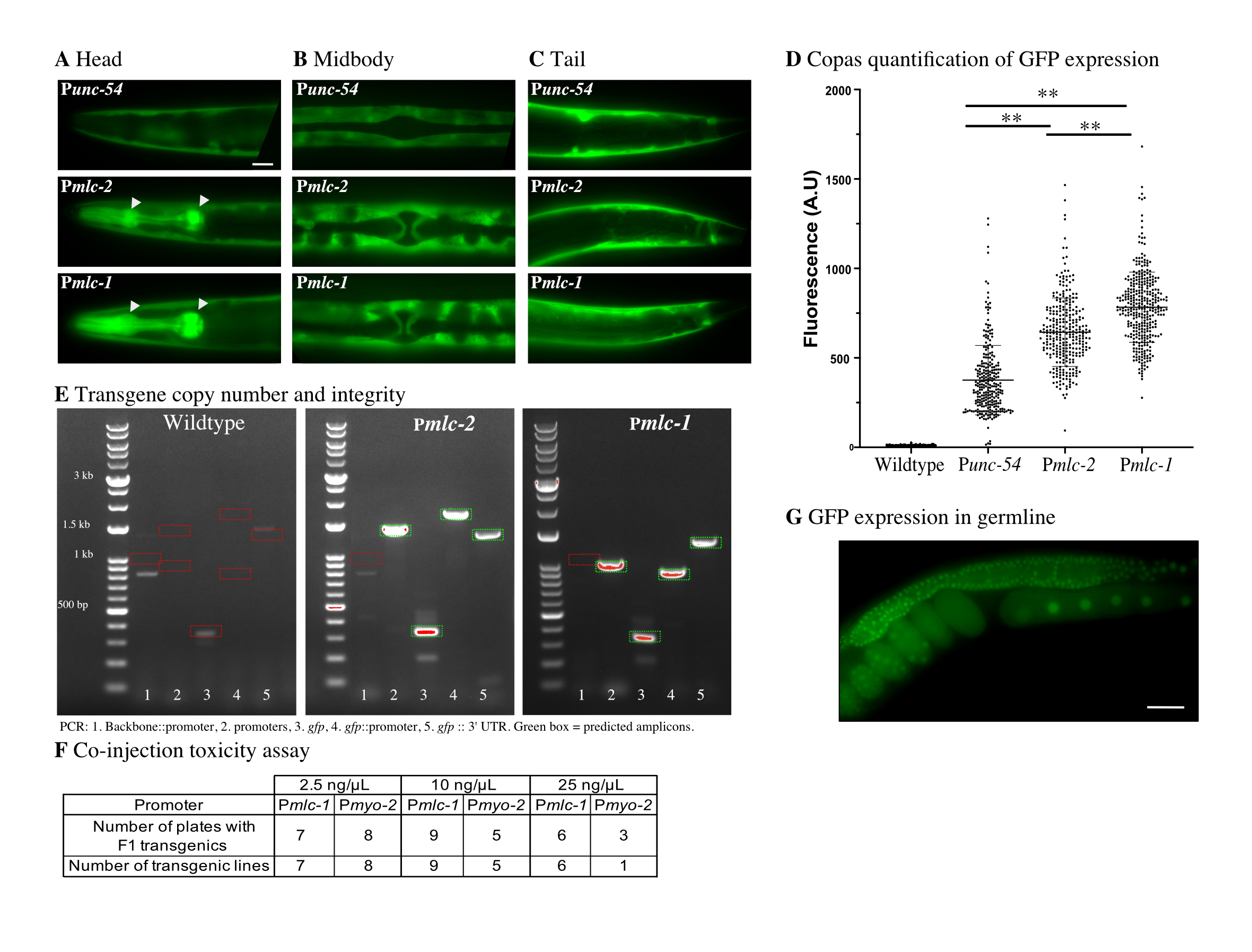
We focused on promoters from genes expected to have muscle-specific expression and manually checked their relative expression levels based on RNA-seq from the ModENCODE project (Gerstein et al. 2010). We identified two putative muscle-specific myosin light chain genes, mlc-1 and mlc-2, that are highly expressed from a single divergent locus. RNA expression levels based on Fragments Per Kilobase Million reads (FPKM) are comparatively high for these two genes in young adults, with 500 FPKM (mlc-1) and 1159 FPKM (mlc-2) compared to 230 FPKM (unc-54), 14 FPKM (myo-2), and 29 FPKM (myo-3). We selected unc-54, mlc-1, and mlc-2 as candidates and amplified 1.9 kb, 1.3 kb, and 1.2 kb promoters, respectively. Due to an early splice site (immediately after the start codon), Pmlc-1 also included the first six amino acids of mlc-1. To quantify the relative expression level of the three promoters, we generated otherwise identical transgenes with a codon-optimized gfp under each promoter’s control and made single-copy insertions at the ttTi5605 site using MosSCI. All promoters drove GFP expression in the body wall, stomato-intestinal, and anal depressor muscles (Figure 1A-C). However, only Punc-54 was not expressed in pharyngeal (Figure 1A) and vulval muscles (Figure 1B) in agreement with previous promoter analysis (Okkema et al. 1993). Using a Copas large-particle flow cytometer, we quantified GFP expression in young adults from a mixed population of the MosSCI strains and found that Pmlc-1:gfp resulted in the highest expression, with approximately two-fold more expression than Punc-54 (Figure 1D). The fluorescence signal matched our subjective visual impression from transgenic animals carrying extra-chromosomal arrays. We verified that the Pmlc-1 and Pmlc-2 strains harbored only single transgene insertions (Figure 1E), excluding the possibility that transgene copy-number caused the observed difference in fluorescence.
Co-injection markers are incorporated into extra-chromosomal arrays in proportion to their concentration in the injection mix (Mello et al. 1991). Higher concentrations thus make it less likely to generate array animals lacking the fluorescent marker, which is essential to avoid false positives (i.e., rescued animals with no single-copy transgene inserted). Therefore, we quantified the toxicity of Pmlc-1::mCherry and Pmyo-2::mCherry transgenes when used as co-injection markers at increasing concentrations (2.5 ng/ul, 10 ng/ul, and 25 ng/ul). We quantified the number of injected P0 animals that gave F1 transgenic animals and stable F2 transgenic lines (Figure 1F). We did not observe apparent toxicity from either promoter at low concentrations (2.5 ng/ul and 10 ng/ul). At high concentration (25 ng/ul), we observed a reduction in plates with rescued F1s and fewer stable lines for Pmyo-2, suggesting some toxicity. In contrast, even at high concentrations, Pmlc-1 did not show any apparent toxicity.
Our goal was to use a single, bright co-injection marker to generate MosSCI insertions, and the lack of toxicity allowed us to test Pmlc-1 at high concentration (25 ng/ul). We inserted a germline-expressed Psmu-1::gfp transgene (Spike et al. 2001) by MosSCI using cbr-unc-119 as the positive selection and a Pmlc-1::tagRFP-T transgene as the visual negative selection marker (in addition to Phsp-16.41:peel-1). We injected ten animals and recovered six plates with extra-chromosomal arrays in the F2 or F3 generation. We heat-shocked these plates and identified four plates with rescued animals (i.e., animals with normal movement) but with no visible red fluorescence on a dissection fluorescence microscope. To rule out false positives, we picked a single worm from each plate and checked for stable germline expression from a transgene insert in the next generation (Figure 1G). All four independent lines expressed GFP in the germline and we observed no tagRFP expression from the fluorescent co-injection marker at high magnification. These results indicate that we could use a single fluorescence marker to select against false positives. Homologous repair relies on unique sequences in the extra-chromosomal array. Therefore, we have generated a set of green (gfp and mNeonGreen) and red (mCherry and TagRFP-T) Pmlc-1 fluorescent markers with cytoplasmic and nuclear expression to avoid cross-talk with the repair machinery.
Here, we describe two promoters from mlc-1 and mlc-2 that can drive bright fluorescent expression in all muscles. We have used mlc-1 to generate co-injection markers that are non-toxic at high concentrations. The use of a single fluorescent co-injection marker solves a minor inconvenience when generating MosSCI injection mixes and would, presumably, also be useful for other methods that rely on similar selection schemes. More generally, mlc-1 and mlc-2 promoters should be generally useful when a bright pan-muscle expression is desired, e.g., for transgene rescue, for expressing optogenetic effectors (e.g., Kerr et al. 2000), or for isolating cells or nuclei from all muscles (e.g., Serizay et al. 2020). Pmlc-2 is expressed at slightly lower levels but may be preferable in the cases where the first six amino acids of mlc-1 might perturb experiments. To facilitate the use of these reagents, we have deposited fluorescent transgenes and Gateway-compatible promoter vectors at Addgene for distribution.
Methods
Request a detailed protocolReagents
Co-injection markers available at Addgene
pSEM228 – Pmlc-1::mNeonGreen(2xNLS)::cbr-tbb-2 UTR (Addgene #159895)
pSEM229 – Pmlc-1::mNeonGreen::cbr-tbb-2 UTR (Addgene #159896)
pSEM230 – Pmlc-1::GFP(2xNLS):: cbr-tbb-2 UTR (Addgene # 159794)
pSEM231 – Pmlc-1::GFP::cbr-tbb-2 UTR (Addgene #159897)
pSEM232 – Pmlc-1::TagRFP-T(2xNLS)::cbr-tbb-2 UTR (Addgene #159898)
pSEM233 – Pmlc-1::TagRFP-T::cbr-tbb-2 UTR (Addgene #159899)
pSEM234 – Pmlc-1::mCherry(2xNLS)::cbr-tbb-2 UTR (Addgene # 159795)
pSEM235 – Pmlc-1::mCherry::cbr-tbb-2 UTR (Addgene #159900)
pCFJ2113 – [4-1] – Promoter – Pmlc-1 (w ATG) (Addgene #159888)
pSEM223 – [4-1] – Promoter – Pmlc-2 (no start) (Addgene #159889)
Annotated plasmid sequences are available at www.wormbuilder.org and https://www.addgene.org/Christian_Froekjaer-Jensen/
Molecular biology
We generated all co-injection markers by three-fragment multisite Gateway reactions (Invitrogen Cat #12538200). The pENTR[4-1] vectors were made by amplifying the promoters and assembled by BP Gateway reaction (Invitrogen Cat #11789013) into a custom pDONR4-1 vector (pCFJ1509). All PCRs were done using high fidelity Phusion polymerase (F530S) and validated by Sanger sequencing.
Promoter quantification
Transgenes with Pmlc-1, Pmlc-2, or Punc-54 fused to a codon-optimized gfp were inserted by MosSCI into the EG4322 (ttTi5605 ; unc-119(ed3)) strain following standard protocols. The injection mixes were composed of 25 ng/ul repair template, 20 ng/ul pCFJ104 (Pmyo-3::mCherry), 20 ng/ul pGH8 (Prab-3::mCherry), 5 ng/ul pCFJ90 (Pmyo-2::mCherry), 20 ng/ul pMA122 (peel-1::HS), 20 ng/ul pNP403 (histamine selection), 20 ng/ul pCFJ1532 (Psmu-1:Mos1 transposase). After injection, we singled each worm onto a standard NGM plate seeded with HB101 and placed animals at 25º Celsius. Starved plates with unc-119 rescued animals were placed for one hour at 37º Celsius (heat-shock) and “chunked” to a new NGM plate. Twenty-four hours later, we picked a single worm with unc-119 rescue and without visible fluorescence from co-injection markers to generate a stable line. Once a line was generated, we placed worms onto standard NGM plates seeded with OP50 and grew at 25º Celsius.
Plasmids used to generate MosSCI insertion strains
pSEM218 – Punc-54::gfp::cbr-tbb-2 UTR
pSEM219 – Pmlc-2::gfp::cbr-tbb-2 UTR
pSEM220 – Pmlc-1::gfp::cbr-tbb-2 UTR
MosSCI strains
CFJ62: kstSi26[Punc-54 | gfp | cbr-tbb-2 3′ UTR] II; unc-119(ed3) III
CFJ63: kstSi27[Pmlc-2 | gfp | cbr-tbb-2 3′ UTR] II; unc-119(ed3) III
CFJ64: kstSi28[Pmlc-1 | gfp | cbr-tbb-2 3′ UTR] II; unc-119(ed3) III
To test if a single co-injection marker was sufficient to select for MosSCI insertions, we used the same strategy but with a germline-expressed transgene and a modified injection mix. The mix consisted of 25 ng/ul pCFJ1805 (Psmu-1::gfp) for germline expression, 25 ng/ul pSEM233 (Pmlc-1::TagRFP-T), 20 ng/ul pMA122 (peel-1 ::HS), 20 ng/ul pCFJ1532 (Psmu-1:Mos1 transposase), GeneRuler 1kb plus DNA ladder (ThermoFisher SM1331) for a final concentration of 100 ng/ul.
Fluorescence quantification
We placed six worms onto NGM plate seeded with OP50 (four plates per strain) and placed animals at 25º Celsius. After five days, worms were washed off growth plates with M9 and washed twice to remove bacteria. We quantified fluorescence using a Copas flow cytometer (Union Biometrica). A mixed population was analyzed, but we only quantified the fluorescence of young adult worms (based on “time of flight,” i.e., animal length).
Transgene toxicity analysis
Transgenic worms were generated by injection into N2 (wild type) worms. The injection mixes were composed of Pmlc-1 or Pmyo-2 fused to mCherry at different concentrations (2.5 ng/ul ; 10 ng/ul ; 25 ng/ul) and GeneRuler 1kb plus DNA ladder (ThermoFisher SM1331) for a final concentration of 100 ng/ul. Ten worms were injected per condition, singled onto NGM plates seeded with OP50, and placed at 25º Celsius. 48 hours after, we determined the number of plates with transgenic F1s and several days later determined if the plates had stable arrays lines in the F2 or F3 generation.
Plasmids used for toxicity assays
pSEM235 – Pmlc-1::mCherry
pCFJ90 – Pmyo-2::mCherry
Microscopy
We imaged animals on a Leica DM2500 microscope equipped with a Leica DFC7000 GT camera at 20X. Worms were immobilized using an M9 solution containing 50 mM of sodium-azide and mounted on 2% agarose pads. We analyzed the fluorescence signal using ImageJ and Affinity designer software.
References
Funding
KAUST intramural funding
Reviewed By
Daniel J DickinsonHistory
Received: August 24, 2020Revision received: August 30, 2020
Accepted: September 1, 2020
Published: September 3, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
El Mouridi, S; Peng, Y; Frøkjær-Jensen, C (2020). Characterizing a strong pan-muscular promoter (Pmlc-1) as a fluorescent co-injection marker to select for single-copy insertions. microPublication Biology. 10.17912/micropub.biology.000302.Download: RIS BibTeX