Interdisciplinary Biological Sciences Program, Northwestern University, Evanston, IL 60208
Robert H. Lurie Comprehensive Cancer Center, Northwestern University, Chicago, IL 60611
Description
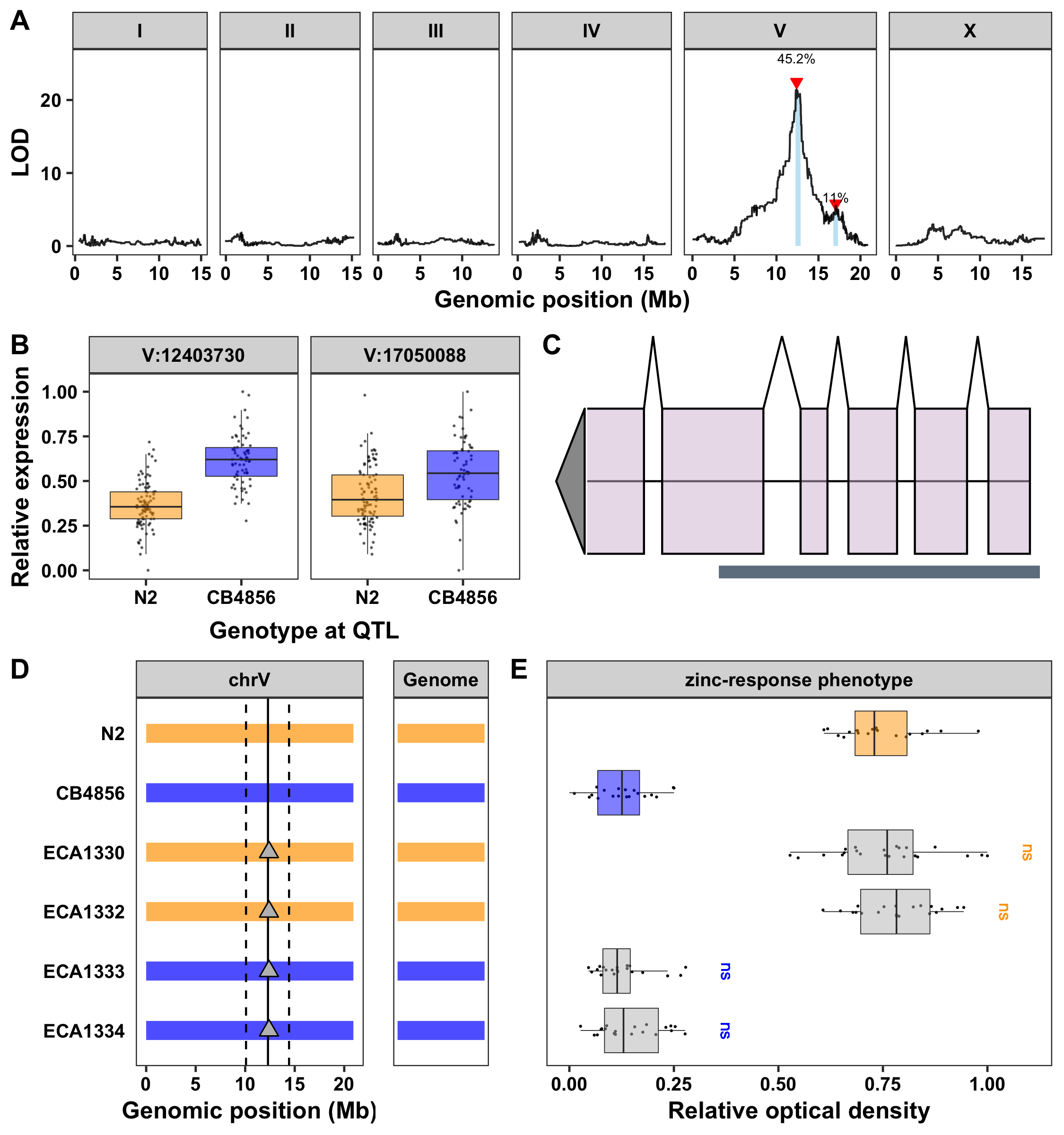
Regulation of the essential trace element zinc is necessary to avoid the toxic consequences caused by too little or too much of this metal (Vallee and Falchuk 1993; Rosen 2006). The zinc-response pathway has been extensively studied in the nematode roundworm Caenorhabditis elegans and several genes have been discovered that function to modulate sensitivity to both high and low zinc concentrations (Dietrich et al. 2016). Recently, we identified a quantitative trait locus (QTL) on the center of chromosome V, indicating that natural genetic variation between the laboratory strain, N2, and a genetically divergent wild isolate from Hawaii, CB4856, contributes to differential responses to excess zinc (Evans et al. 2020).
The confidence interval for this QTL is 1.6 Mb and contains 629 genes (WS263). Of these genes, 113 have one or more genetic variants predicted to modify the amino-acid sequence of the protein. However, protein-coding variation is just one of the ways that genetic variation can cause phenotypic variation. Another is variation in gene expression, which is hypothesized to be important in the majority of complex traits (Hindorff et al. 2009). We identified 83 expression QTL (eQTL) within this 1.6 Mb region using the eQTL dataset that mapped expression differences among a panel of recombinant inbred advanced intercross lines (RIAILs) also derived from N2 and CB4856 (Rockman et al. 2010; Evans and Andersen 2020). The most significant eQTL in this region caused a change in expression of the gene cdr-6 (Figure 1A). This gene, a homolog of the cadmium-response gene cdr-1, is highly expressed in intestinal and pharynx muscle cells during larval stages and is downregulated after treatments with arsenic, cadmium, or zinc (Dong et al. 2008). Furthermore, cdr-1 was previously shown to mitigate cadmium toxicity in C. elegans (Dong et al. 2005; Hall et al. 2012). Together, these data suggest that expression of cdr-6 might be toxic to C. elegans in the presence of heavy metals. Additionally, RIAILs with the CB4856 allele on chromosome V naturally express higher levels of cdr-6 (Figure 1B) and are also more sensitive to zinc than RIAILs with the N2 allele (Evans et al. 2020). This result indicates that strains with naturally low expression of cdr-6 are more resistant to excess zinc than strains with naturally high expression of this gene.
To test this hypothesis, we used CRISPR-Cas9 genome editing to create strains with large deletions of cdr-6 in both the N2 and CB4856 genetic backgrounds (Figure 1C,D). Because expression of cdr-6 was higher in RIAILs with the CB4856 allele (associated with zinc sensitivity) than in RIAILs with the N2 allele (associated with zinc resistance) (Figure 1B), we expected that a knockout of cdr-6 in the CB4856 genetic background would cause increased resistance to excess zinc. Alternatively, if variation in expression of cdr-6 underlies the zinc-response QTL on chromosome V, a knockout of cdr-6 in the N2 genetic background should not cause an increase in zinc resistance. We exposed N2, CB4856, and two strains with independently derived cdr-6 deletion alleles in each genetic background to elevated zinc and measured their optical densities using a high-throughput assay with the COPAS BIOSORT (Andersen et al. 2015; Evans and Andersen 2020; Evans et al. 2020). We found that strains with a deletion of cdr-6 phenocopied the strain with the same genetic background (Figure 1E), suggesting that differences in expression of cdr-6 do not underlie zinc responses.
This study not only provides evidence against cdr-6 as the causal gene underlying differences in zinc resistance between the N2 and CB4856 strains, but also indicates that cdr-6 does not influence zinc resistance in C. elegans. Although a previous study showed that cdr-6 was downregulated in response to zinc (Dong et al. 2008), our results do not contradict theirs. In fact, the authors also showed that the accumulation of fluid-filled droplets in the pseudocoelom, a phenotype observed in strains with inhibited CDR-6 expression using RNAi, is not increased in response to zinc or cadmium exposure (Dong et al. 2008). Taken together, we conclude that expression of cdr-6 decreases in response to zinc but animal development in the presence of zinc is not affected by cdr-6 function. It is likely that cdr-6 does not function in the nematode zinc response but rather is downregulated as an indirect effect of zinc exposure.
Methods
Request a detailed protocolStrains
Animals were grown at 20°C on modified nematode growth media (NGMA) containing 1% agar and 0.7% agarose to prevent burrowing and fed OP50 (Andersen et al. 2014). The cdr-6 deletion strains are described below. All strains are available upon request.
| Strain | Genotype | Deletion |
| ECA1330 | cdr-6(ean181)
[N2 background] |
730 bp deletion from V:12,413,681-12,412,951 |
| ECA1332 | cdr-6(ean183)
[N2 background] |
811 bp deletion from V:12,413,723-12,412,912 |
| ECA1333 | cdr-6(ean184)
[CB4856 background] |
632 bp deletion from V:12,413,034-12,413,665 |
| ECA1334 | cdr-6(ean185)
[CB4856 background] |
788 bp deletion from V:12,412,864-12,413,651 |
Expression QTL mapping
Microarray expression for 14,107 probes were previously collected from 208 RIAILs (Rockman and Kruglyak 2009; Rockman et al. 2010), filtered (Andersen et al. 2014), and mapped using linkage mapping with 13,003 SNPs (Brady et al. 2019; Evans and Andersen 2020). The probe that measures expression for cdr-6 is A_12_P101644. All expression and eQTL data can be found in Evans and Anderson, 2020.
Generation of cdr-6 deletion strains
Deletion alleles for cdr-6 were generated using CRISPR-Cas9 genome editing as previously described (Hahnel et al. 2018; Brady et al. 2019). Briefly, 5’ and 3’ guide RNAs (Synthego, Redwood City, CA) were designed with the highest possible on-target and off-target scores (Doench et al. 2016). The CRISPR injection mix (1 µM dpy-10 sgRNA, 5 µM of each sgRNA for cdr-6, 0.5 µM of a single-stranded oligodeoxynucleotide template for homology-directed repair of dpy-10 (IDT, Skokie, IL), and 5 µM Cas9 protein (Q3B Berkeley, Berkeley, CA) in water) was assembled and incubated for an hour before injection. Young adults were mounted onto agar injection pads, injected in either the anterior or posterior arm of the gonad, and allowed to recover on 6 cm plates. Survivors were transferred to individual 6 cm plates 12 hours later and allowed to lay embryos. After two to three days, the F1 progeny were screened and individuals with Rol or Dpy phenotypes were selected and transferred to new 6 cm plates. After 48 hours, the F1 individuals were genotyped by PCR flanking the desired deletions. Individuals with deletions were propagated and genotyped for at least two additional generations to ensure homozygosity and to cross out the Rol mutation. Deletion amplicons were Sanger sequenced to identify the exact location of the deletion. Reagents used to generate and validate the deletion are found below:
crECA36 dpy-10 guide RNA:
GCUACCAUAGGCACCACGAG
crECA37 dpy-10 repair construct: CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTA GGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT
cdr-6 guide RNA:
crECA153: TCCAGTGACAACAACCCTAG
crECA115: AGTTTATAGTCGTTGTGTTG
External primers for deletion validation:
oECA1635: ACACACGTTCACTGGCTAGACT
oECA1636: AGCACGTCGTTGATATGCGAAC
Internal primers for deletion validation:
oECA1365: ACACACGTTCACTGGCTAGACT
oECA1637: TGTGTTCCCCATTGAGCTCGAT
High-throughput zinc-response assay
Zinc response was measured as a function of animal optical density (representing nematode development) as described previously (Brady et al. 2019; Evans and Andersen 2020). Briefly, strains were propagated for two generations, bleach-synchronized, and titered in K medium (Boyd et al. 2012) at a concentration of 25-50 embryos per well of a 96-well microtiter plate. The following day, arrested L1s were fed HB101 bacterial lysate (Pennsylvania State University Shared Fermentation Facility, State College, PA; (García-González et al. 2017)) at a final concentration of 5 mg/mL with either 1% water or 250 µM zinc in 1% water. Nematodes were allowed to grow at 20°C with constant shaking for 48 hours, treated with sodium azide (5 mM in water), and analyzed using the COPAS BIOSORT (Union Biometrica; Holliston, MA). Animal optical densities collected by the BIOSORT were processed and analyzed using the R package easysorter (Shimko and Andersen 2014) as described previously (Brady et al. 2019). Each strain has a minimum of 20 replicates and each replicate is defined by the median animal optical densities of six to 32 animals. Pairwise tests for statistically significant differences in the zinc response between strains were performed using the TukeyHSD function (R Core Team 2017) on an ANOVA model with the formula (phenotype ~ strain). For plotting purposes, these residual values were normalized from zero to one with zero being the well with the smallest value and one the well with the largest value.
Acknowledgments
We would like to thank members of the Andersen laboratory for their helpful comments on the manuscript. We would also like to thank WormBase and the C. elegans Natural Diversity Resource for data critical for our analysis.
References
Funding
This work was supported by R01 ES029930 from the National Institute of Environmental Health Sciences to E.C.A. Additionally, K.S.E. received support from the NSF-Simons Center for Quantitative Biology at Northwestern University (awards Simons Foundation/SFARI 597491-RWC and the National Science Foundation 1764421) and the Cell and Molecular Basis of Disease Training grant (T32-GM008061).
Reviewed By
Joel MeyerHistory
Received: August 2, 2020Revision received: September 4, 2020
Accepted: September 5, 2020
Published: September 14, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Evans, KS; Andersen, EC (2020). The cadmium-responsive gene, cdr-6, does not influence Caenorhabditis elegans responses to exogenous zinc. microPublication Biology. 10.17912/micropub.biology.000305.Download: RIS BibTeX