Department of Psychology, University of British Columbia, 2136 West Mall, Vancouver, British Columbia V6T 1Z4, Canada
Description
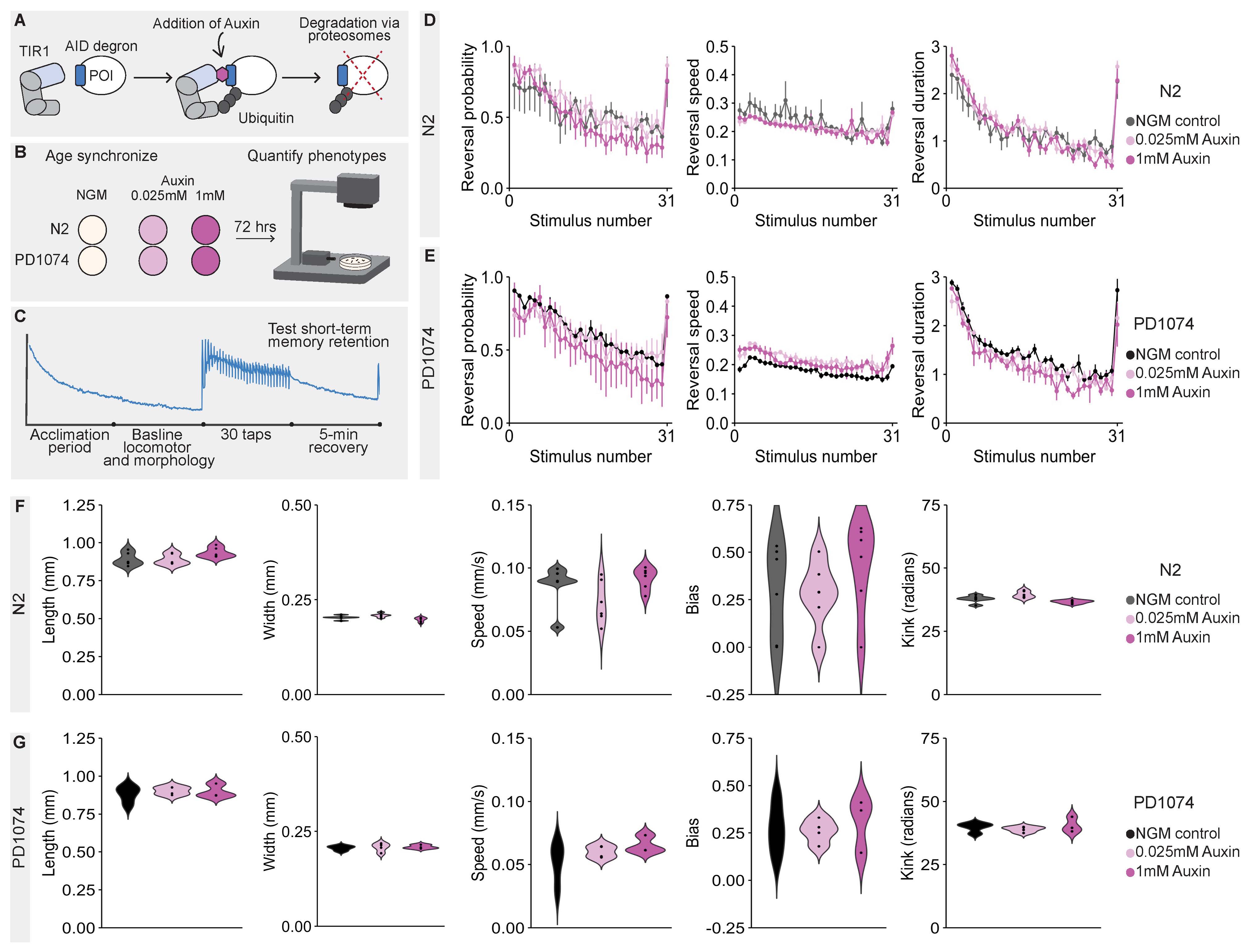
The Auxin-Inducible Degradation (AID) system is a powerful technique in the C. elegans toolkit that enables conditional and reversible protein depletion with high temporal and spatial specificity (Zhang et al. 2015; Martinez et al. 2020; Ashley et al. 2020; Martinez and Matus 2020). This system relies on tagging a gene of interest with a short AID degron sequence and transgenic expression of TIR1, an inducible E3 ubiquitin ligase normally found only in plants (Nishimura et al. 2009; Zhang et al. 2015). Upon exposure to the plant-derived hormone Auxin, TIR1 is activated and targets AID-tagged proteins for proteasomal degradation (Nishimura et al. 2009; Zhang et al. 2015) (Figure 1A). While there are qualitative reports that Auxin does not overtly affect the morphology or behavior of wild-type C. elegans (Zhang et al. 2015), this has not been quantitatively assessed. Determining whether Auxin significantly affects C. elegans morphology and behavior, even in subtle ways, is important given the C. elegans community’s rapid uptake of the AID system (Kasimatis et al. 2018; Nance and Frøkjær-Jensen 2019; Ashley et al. 2020; McDiarmid et al. 2020). Here, we use our high-throughput machine vision tracking system, the Multi-Worm Tracker (MWT) (Swierczek et al. 2011), to investigate whether exposure to Auxin affects a suite of morphological, locomotor, mechanosensory, and short-term habituation learning phenotypes in our lab’s derivative of Bristol N2 wild-type worms and the CGC wild-type reference strain, PD1074 (Yoshimura et al. 2019) (Figure 1B).
We report that lifelong exposure to 0.025mM or 1mM Auxin does not significantly affect any of the objectively quantified phenotypic features in either N2 or PD1074 wild-type strains (Figure 1D-G). We tested 1mM Auxin as it is commonly used to quickly and fully degrade AID tagged proteins, whereas, in our experience, 0.025mM Auxin still enables protein degradation but is dilute enough to allow AID tagged proteins to be re-expressed within 24 hours (hrs) of the animals being off Auxin. The Auxin plates used were indeed effective, as plates from the same batch were able to induce lethality in the CA1210 (dhc-1::AID::GFP) positive control strain (this is expected due to ubiquitous degradation of the essential dynein heavy chain DHC-1) (Zhang et al. 2015). An additional concern has been that Auxin is dissolved in ethanol (Martinez et al. 2020), and, as many of us know first-hand, ethanol can indeed alter animal behavior (Mitchell et al. 2007; Spear 2018). Importantly, we find that the ethanol used to dissolve Auxin does not affect these phenotypes when tested on agar plates, likely because the ethanol evaporates during the 72 hrs plates are allowed to dry. NOTE: The ethanol that Auxin is dissolved in will likely affect these phenotypes if using liquid culture (not tested here).
We hope this article will serve as a resource to show that with proper preparation, Auxin can be used with the AID system for effective protein depletion in C. elegans with minimal effects on a suite of morphological and behavioral phenotypes studied by the community. However, we still recommend the inclusion of a wild-type on Auxin negative control for behavioral experiments to ensure there is nothing wrong with the Auxin solution that may affect behavior (e.g. contamination).
Methods
Request a detailed protocolAuxin administration
Auxin treatment was performed by transferring animals to bacteria-seeded NGM plates containing Auxin. A 400 mM stock solution of Auxin indole-3- acetic acid (IAA) (Thermo Fisher, Alfa Aesar™ #A1055614) was created by dissolving Auxin in ethanol. The Auxin stock was then diluted into separate flasks of molten NGM agar to final concentrations of 0.025mM and 1mM (molten NGM was cooled to ~50°C before the addition of Auxin), and were poured into Petri plates. Auxin plates were allowed 72 hrs to dry in the dark, and were then seeded with 50 µl of OP50 liquid culture 48 hrs before use. Animals were continuously exposed to Auxin as they were age synchronized (as described below) onto Auxin plates and tested at 72 hrs post-lay.
Behavioral assays
Animals were age synchronized onto NGM and Auxin Petri plates which were seeded with 50 µl of OP50 liquid culture and dried in the dark for 72 hrs. Four to 6 plates were synchronized for each condition by picking five gravid adults onto each plate and allowing the animals to lay eggs for 4 hrs before removal (resulting in 50-100 worms per plate) (Figure 1B). Animals were then maintained in the dark in a 20°C incubator for 72 hrs.
The behavioral paradigm (Figure 1C) consisted of a 5-minute (min) acclimation period followed by a 5-min period to assess morphological and baseline locomotion phenotypes. Thirty mechanosensory stimuli were then administered to the side of the Petri plate by the MWT’s automated piston at a 10 second (sec) interval. In response to the mechanosensory stimuli, animals respond by briefly crawling backwards then resuming forward locomotion (i.e. a reversal response) (Rankin et al. 1990), allowing multiple features of response sensitivity and habituation to be assessed (Swierczek et al. 2011; McDiarmid et al. 2020). Following the 30th stimulus, animals were allowed to recover for 5 mins. A final stimulus was then administered to test spontaneous recovery from short-term habituation (short-term memory retention).
Multi-Worm Tracker behavioral analysis and statistics
We used the MWT software (version 1.2.0.2) for stimulus delivery and image acquisition (Swierczek et al. 2011), and Choreography software (version 1.3.0_r103552) for phenotypic quantification. The filters, “–shadowless”, “–minimum-move-body 2”, and “–minimum-time 20” were used to identify animals that moved ≥ 2 body lengths and were tracked for ≥ 20 secs. Morphology and baseline locomotion features were identified using standard choreography output commands (Swierczek et al. 2011). Reversals that occurred within 1 sec after the administration of a mechanosensory stimulus were identified using the MeasureReversal plugin (Swierczek et al. 2011). Comparisons of “final response” comprised the average of the final three stimuli. For a complete description of the 26 phenotypic features, see the MWT user guide (https://sourceforge.net/projects/mwt/). All data is freely available upon request. Custom R scripts organized and summarized Choreography output files which are freely available at https://github.com/troymcdiarmid/MWT_Wildtype_Auxin/releases/tag/v1.0. (An archived version of these data and scripts are available on Caltech Data https://doi.org/10.22002/d1.1639). Condition blinding was not necessary as the MWT scores behavior objectively (Swierczek et al. 2011). Phenotypic features were pooled across the 4-6 plate replicates (50-100 animals per plate) for each strain and means of each condition were compared with an unpaired t-test and Benjamini-Hochberg control of the false discovery rate at 0.01. Final figures were generated using the ggplot2 package in R (Wickham 2016). Images taken by the MWT were used to confirm lethality of CA1210 (dhc-1::AID::GFP) animals after 72 hrs of Auxin exposure.
Reagents
N2 wild-type, PD1074 wild-type, and CA1210 dhc-1(ie28[dhc-1::degron::GFP]) I; ieSi57 II. All strains are available from the CGC.
References
Funding
This project was supported by a Canadian Institutes of Health Research (CIHR) Doctoral Research Award to T.A.M.; a Natural Sciences and Engineering Research Council (NSERC) Master’s Research Award to L.D.K; and a CIHR project grant (CIHR MOP PJT-165947) to C.H.R.
Reviewed By
AnonymousHistory
Received: September 4, 2020Revision received: September 26, 2020
Accepted: September 30, 2020
Published: October 8, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
McDiarmid, TA; Kepler, LD; Rankin, CH (2020). Auxin does not affect a suite of morphological or behavioral phenotypes in two wild-type C. elegans strains. microPublication Biology. 10.17912/micropub.biology.000307.Download: RIS BibTeX