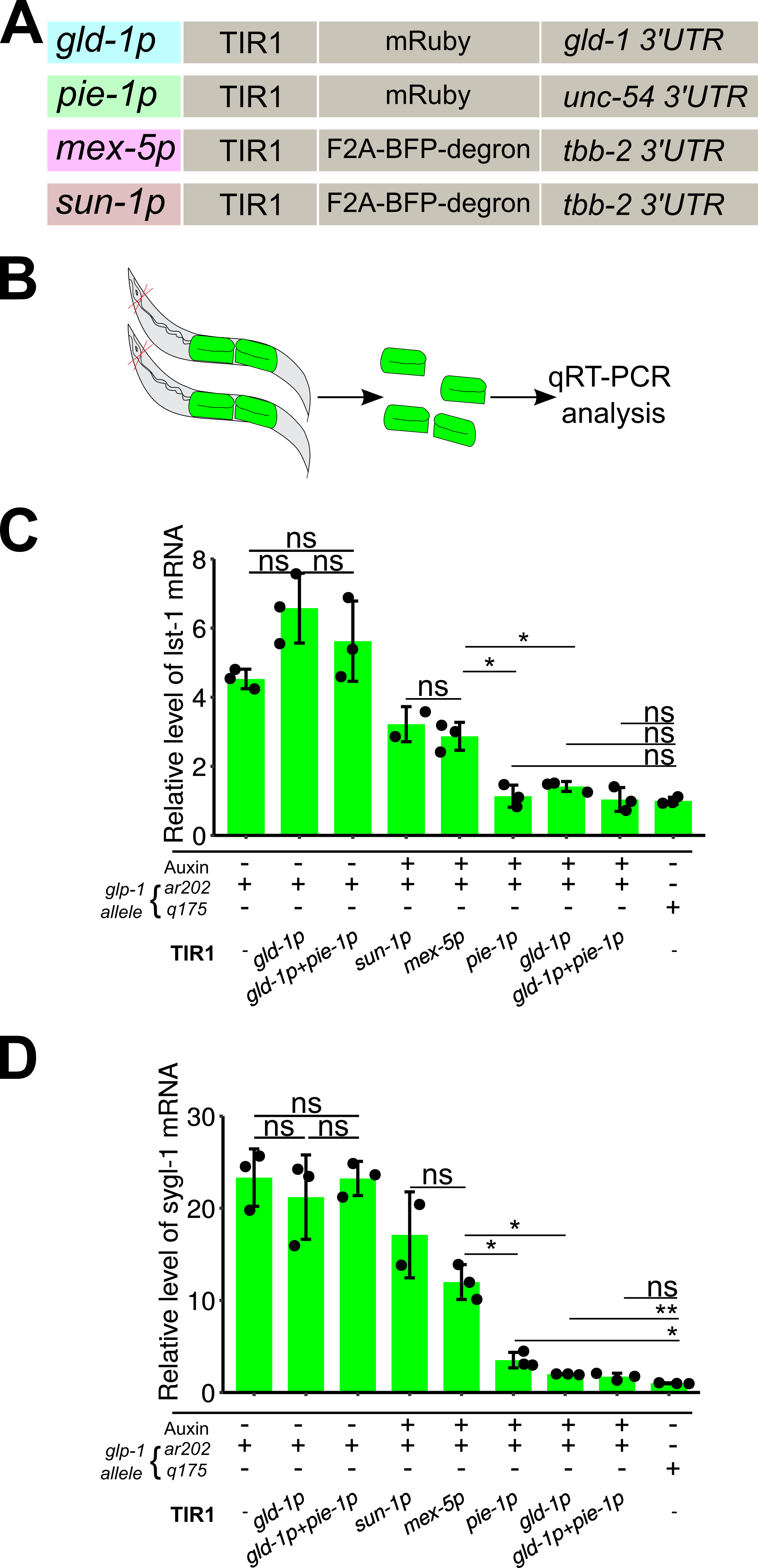
Description
The auxin inducible degradation (AID) system was first introduced in C. elegans by Dernburg lab and has become a widely-used approach to study tissue-specific and/or temporal aspects of gene function (Zhang et al. 2015; Ashley et al. 2020; Martinez et al. 2020). The AID system utilized a plant derived E3 ubiquitin ligase, TIR1, to specifically degrade proteins fused with the “degron” tag following treatment with the plant growth hormone auxin. To increase the utility of the AID system, the Ward laboratory recently generated an expanded set of TIR1s transgenes, controlled by different tissue-specific promoters (Ashley et al. 2020). Here we aim to compare different germline-expressed TIR1 transgenes for their efficiency in degrading the transcription factor LAG-1, which is C-terminally tagged with degron (Chen et al., 2020). As indicated in Figure 1, these TIR1 transgenes are driven by the following promoters: gld-1p (Zhang et al. 2015), mex-5p (Ashley et al. 2020), sun-1p (Ashley et al. 2020), and pie-1p (Kasimatis et al. 2018), and contain the indicated C-terminal fluorescent proteins and 3’ untranslated regions (Fig. 1A).
In the C. elegans germline, GLP-1 Notch signaling employs transcriptional targets, lst-1 and sygl-1, to maintain the germline stem cell (GSC) fate (Kershner et al. 2014; Chen et al. 2020). In the nucleus, the transcription factor LAG-1 and GLP-1(INTRA) form a complex that activates transcription of targets (Lee et al., 2016; Chen et al., 2020). LAG-1 is widely expressed and is also required for LIN-12 Notch signaling in somatic cells (Greenwald and Kovall 2013). Utilizing the AID system, we recently demonstrated that LAG-1 functions germline-autonomously to promote the GSC fate (Chen et al. 2020).
We have previously shown that the LAG-1 protein levels are reduced to background using the gld-1p::TIR1 transgene (Chen et al. 2020). However, the low level of endogenous LAG-1 in the germline stem cell region, with or without auxin treatment, makes quantitative protein level comparisons challenging. Here, we measured the level of mRNA for the LAG-1 transcriptional targets lst-1 and sygl-1 by quantitative real-time PCR (qRT-PCR) from dissected gonads (Fig. 1B), as an indirect but more sensitive output for measuring degradation efficiency of LAG-1::degron by different TIR1 transgenes. Since LAG-1 degradation in the wild type germline caused the GSCs to enter meiosis, the qRT-PCR experiments were conducted in the gld-2 gld-1 double mutant background (see complete genotype in the Reagent section). GLD-1 and GLD-2 are the two major pathways that promote GSCs to enter meiosis. In gld-2 gld-1 double mutants the mitotically cycling germ cells fail to enter meiosis and results in a germline tumor, irrespective of GLP-1 signaling or LAG-1 function (Kershner et al. 2014; Chen et al. 2020). In addition, the glp-1 gain of function allele ar202 was included in the gld-2 gld-1 double mutant background, allowing the transcription of lst-1 and sygl-1 mRNA to occur throughout the germline, thus increasing signal-to-noise ratio. The auxin treatment and qRT-PCR experiments were conducted as previously described (Chen et al. 2020). Starvation synchronized L1 animals were grown on NGM plates supplemented with or without 1 mM auxin for 48 hours at 25 °C. After auxin treatment, ~50 adult animals were dissected to isolate the gonads (Fig. 1B, Chen et al. 2020). The relative mRNA levels of lst-1 and sygl-1 are presented in Figure 1C & D.
One potential issue for the AID system is auxin independent TIR1 degradation of the target proteins. With the lag-1::degron allele, we have previously shown that both LAG-1 protein accumulation and progenitor zone size are largely normal in the presence of gld-1p::TIR1, without auxin (Chen et al. 2020). Here we compared the mRNA levels of lst-1 and sygl-1 from three different strains grown on NGM plates without auxin (Fig. 1C & D). The data showed that both lst-1 and sygl-1 mRNA level were not significantly different among animals without auxin regardless of which TIR transgene they carried (Fig. 1C & D), indicating that auxin independent TIR1 degradation of LAG-1 did not occur. We found that all four TIR1s can degrade LAG-1 sufficiently to generate Glp-1 like sterile animals in otherwise wild type background. In the tumorous germlines, the levels of lst-1 and sygl-1 mRNA were reduced in auxin treated animals, also supporting the conclusion that the AID system is knocking down LAG-1 protein. Importantly, we observed that lst-1 and sygl-1 mRNA level was lower in the gld-1p::TIR1 and pie-1p::TIR1 backgrounds compared to that in the sun-1p::TIR1 and mex-5p::TIR1 backgrounds, after auxin treatment (Fig. 1). When compared to glp-1(q175) null background, we noticed that the average expression level of lst-1 and sygl-1 mRNA was slightly higher in gld-1p::TIR1, pie-1p::TIR1, and gld-1p::TIR1 + pie-1p::TIR1 background after auxin treatment. Statistical analysis indicates the lst-1 mRNA level was indistinguishable compared to glp-1 null germline, while the sygl-1 mRNA level is slightly but significantly higher than that in glp-1 null germlines. Together, we concluded that gld-1p::TIR1 and pie-1p::TIR1 are more efficient in degrading LAG-1 in the germline stem cell region.
It is not known what molecular feature(s) of the TIR1 transgenes determined high efficiency degradation of LAG-1::degron in germline stem cells, as the transgenes differ not only in the promoter, but also in the fluorescent protein tag, and in the 3’ UTR. For example, the mex-5p and sun-1p transgenes contain fluorescent protein BFP with a C-terminal degron tag, separated by a self-cleavable linker F2A, a design that allows a direct functional check of TIR1 activity through loss of BFP fluorescence. In contrast, pie-1p::TIR1 and gld-1p::TIR1 have mRuby fused to TIR1 without the degron motif. It is possible that reduced efficiency of mex-5p and sun-1p transgenes, rather than a consequence of different promoters, was due to competition of TIR1 for BFP::degron versus LAG-1::degron. Finally, we recommend testing multiple TIR1 transgenes to determine which one is optimal for the protein, cell type and timing of interest.
Reagents
The following strains are used in this study:
BS3879:
gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
glp-1(q175)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III)
BS4310:
gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III)
BS5629:
gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
ieSi64 [gld-1p::TIR1::mRuby::gld-1 3’UTR + Cbr-unc-119(+)] (II);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III);
lag-1(oz536oz537[lag-1::degron::3xHA]) (IV)
BS5631:
fsIs1 [pie-1p::TIR1::mRuby::unc-54 3’UTR] gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
ieSi64 [gld-1p::TIR1::mRuby::gld-1 3’UTR + Cbr-unc-119(+)] (II);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III);
lag-1(oz536oz537[lag-1::degron::3xHA]) (IV)
BS5630:
fsIs1 [pie-1p::TIR1::mRuby::unc-54 3’UTR] gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III);
lag-1(oz536oz537[lag-1::degron::3xHA]) (IV)
BS7023:
gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
wrdSi8 [mex-5p::TIR1::F2A::mTagBFP2::degron-NLS::tbb-2 3’UTR] (II);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III);
lag-1(oz536oz537[lag-1::degron::3xHA]) (IV)
BS7024:
gld-2(q497) gld-1(q485)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (I);
cpIs103 [sun-1p::TIR1::F2A::mTagBFP2::degron-NLS::tbb-2 3’UTR] (II);
glp-1(ar202)/hT2::gfp [bli-4(e937) let-?(q782) qIs48] (III);
lag-1(oz536oz537[lag-1::degron::3xHA]) (IV)
Acknowledgments
We thank Swathi Arur, Jordan Ward, Dan Dickinson, Ari Pani and Liangyu Zhang for helpful discussions. We thank the Caenorhabditis Genetics Center, funded by National Institutes of Health Office of Research Infrastructure Programs (P40OD-010440) for strains.
References
Funding
R01 GM-100756 to TS
Reviewed By
AnonymousHistory
Received: July 6, 2020Revision received: September 14, 2020
Accepted: September 15, 2020
Published: September 18, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Chen, J; Mohammad, A; Schedl, T (2020). Comparison of the efficiency of TIR1 transgenes to provoke auxin induced LAG-1 degradation in Caenorhabditis elegans germline stem cells. microPublication Biology. 10.17912/micropub.biology.000310.Download: RIS BibTeX