Description
Caveolins are integral membrane proteins responsible for the formation of caveolae, invaginations of the plasma membrane linked to various disease states (Parton et al. 2020). In C. elegans, there are two caveolin proteins, CAV-1 and CAV-2. The cav-1 gene shares homology with all three mammalian caveolin genes (Tang et al. 1997). C. elegans CAV-1 protein does not appear to form caveolae, but a double-knockout mutant of cav-1 and cav-2 affects egg laying, and knockdown of cav-1 affects locomotion in a dynamin mutant background (Parker et al.. 2007, Kirkham et al. 2008, Sato et al. 2008). Based on exogenous expression, CAV-1::GFP is known to localize to cortical granules and the plasma membrane in oocytes and the early embryo, to the plasma membrane in later embryos, as well as to the neuromuscular system in larvae and adult worms (Sato et al. 2006, Bembenek et al. 2007, Parker et al. 2007).
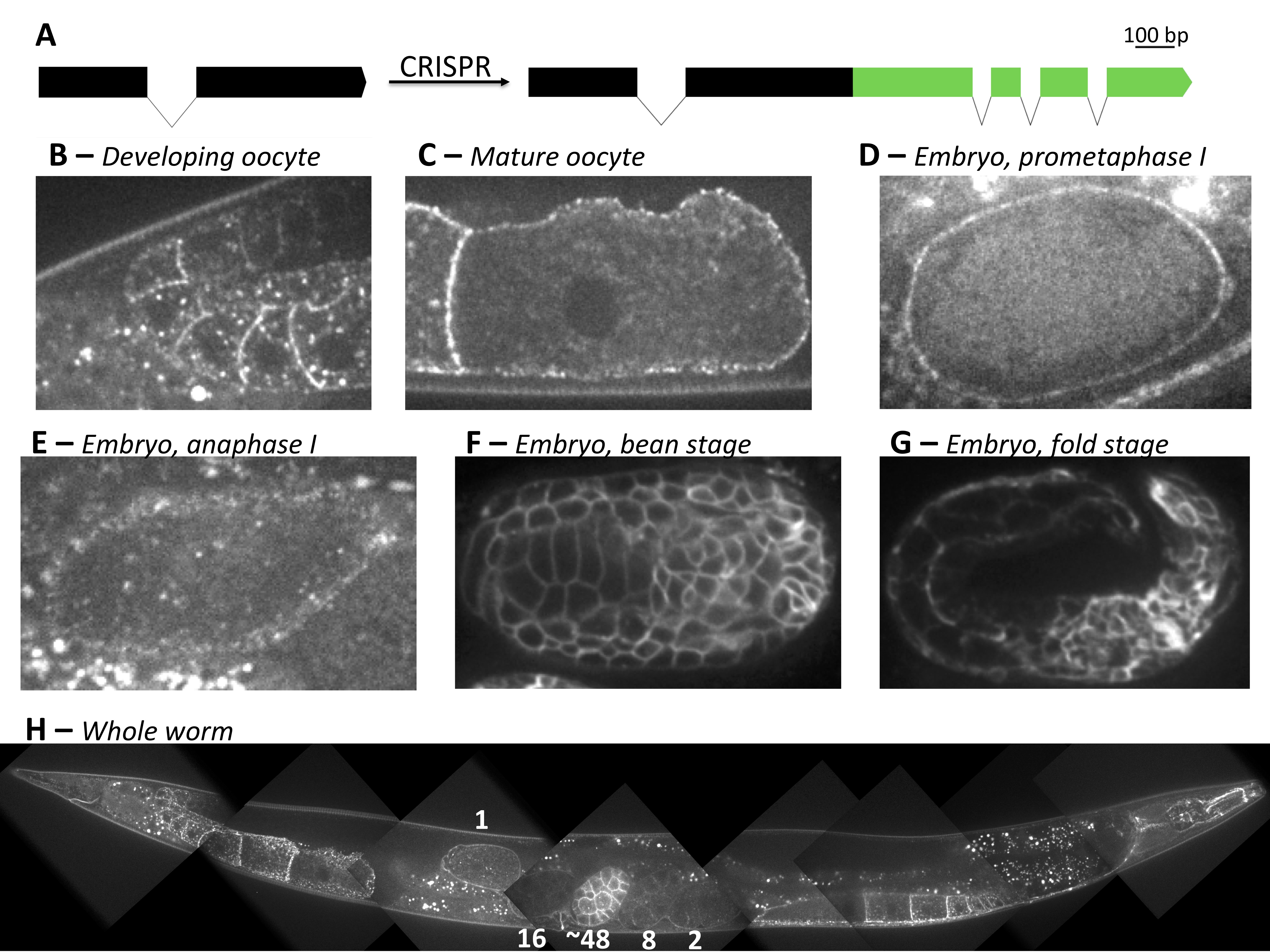
Using CRISPR/Cas9, we generated an endogenous CAV-1::GFP fluorescent fusion strain, primarily to create a cortical granule marker (Figure 1A). Imaging endogenous CAV-1::GFP revealed similar patterns as previously reported in the germ line (Sato et al. 2006). However, unlike the pie-1 driven exogenous line, endogenous CAV-1::GFP was not clearly detected in the distal gonad until the bend region of the gonad arm, where it localized to the plasma membrane, and vesicle-shaped structures (Figure 1B). Oocytes had slightly higher expression showing enrichment at plasma membrane localization but faint cytoplasmic signal, and no obvious enrichment to vesicles (Figure 1C). After ovulation, CAV-1::GFP remained on the plasma membrane during meiosis I and was no longer detected by meiosis II (Figure 1D,E). In 5/5 anaphase I embryos, CAV-1::GFP was not significantly localized to cortical granules at a time when separase could be observed on vesicles. Overall, endogenous CAV-1::GFP signal is less intense in the germline than exogenous pie-1-driven lines. However, endogenous CAV-1::GFP signal became increasingly intense at the plasma membrane sometime after the 16-cell embryo stage (Figure 1H). The membrane signal remains intense through later stages of embryonic development (Figure 1F,G). This potentially indicates a dynamic role for cav-1 in embryonic development. Finally, adult somatic expression appeared to follow previously observed patterns (Figure 1H, Parker et al. 2007).
In summary, we have developed an endogenous CAV-1::GFP line by CRISPR, which can be of use to researchers interested in CAV-1.
Methods
Request a detailed protocolCRISPR/Cas9 Gene editing
We followed the CRISPR/Cas9 protocol generated by Seydoux lab for C-terminal GFP tagging of the C. elegans cav-1 gene (Figure 1A, Paix et al. 2015). The repair template was amplified from the pDD282 plasmid. All guide RNAs and oligos were obtained commercially.
The primer sequences are listed below:
cav-1::GFP Forward (termed DS_027):
ACGGAATCAATCAAGAAACTACTGCTCCATGCGTCATGAGTAAAGGAGAAGAATTGTTCACTG
cav-1::GFP Reverse (termed DS_026):
AGTAAAATGAATTTGAGATAAATTAAATAAATTTACTTGTAGAGCTCGTCCATTCC
The crRNA sequence recognizing the spacer upstream of NGG is as follows:
(Termed DS_g003): AUUAAAUAAAUUUAGACGCA
Microscopy
Imaging was done as indicated in Bai et al. 2020. Embryos were staged using the localization of separase fused with a red fluorescent protein as well as embryo position and age in the animal.
Reagents
JAB192: cav-1(erb78[cav-1::GFP])
This strain is available upon request.
Acknowledgments
We thank members of the Bembenek and Csankovszki labs, especially Chris Turpin, for useful feedback.
References
Funding
National Institutes of Health (R01 GM114471)
Reviewed By
Sara OlsonHistory
Received: August 6, 2020Revision received: September 18, 2020
Accepted: September 18, 2020
Published: September 21, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Sloan, DE; Bembenek, JN (2020). Endogenous expression and localization of CAV-1::GFP in C. elegans. microPublication Biology. 10.17912/micropub.biology.000311.Download: RIS BibTeX