Description
Mechanisms of chemotactic behaviors have been of great interest in C. elegans neuroscience since the early days of its research (Ward 1973). Lewis and Hodgkin (1977) systematically isolated more than ten abnormal chemotaxis (che) mutants that showed defective chemotaxis to sodium (Na+) and chloride (Cl–) ions (Lewis and Hodgkin 1977), whose responsible genes have already been molecularly characterized except for che-5(e1073). We here show that che-5(e1073) is a missense allele of gcy-22, which encodes a receptor guanylyl cyclase (rGC) specifically expressed in the ASE-right (ASER) gustatory neuron and is essential for chemosensation through the neuron.
C. elegans is attracted to the NaCl concentration at which it has experienced with food, while avoid the concentration at which it has experienced starvation. ASER plays a major role in food-associated salt concentration chemotaxis; input of salt information into ASER is required and sufficient for chemotaxis to the salt concentration associated with food (Kunitomo et al. 2013). ASE neurons, consisting of bilaterally symmetrical ASE-left (ASEL) and ASER, are the major sensory neuron for water-soluble attractants (Bargmann and Horvitz 1991). They respectively sense different sets of ions such as Na+ and Cl– (Pierce-Shimomura et al. 2001; Suzuki et al. 2008; Ortiz et al. 2009). A cyclic GMP (cGMP) signaling pathway consisting of rGCs and TAX-4/TAX-2 cyclic nucleotide-gated ion channels mediates sensory transduction in ASE (Coburn and Bargmann 1996; Komatsu et al. 1996; Ortiz et al. 2009). ASEL and ASER express distinct sets of rGCs (Ortiz et al. 2006). Of these, gcy-22 is essential for ASER to respond to multiple ion species; therefore it is proposed as a common component of chemoreceptor complexes (Ortiz et al. 2009; Adachi et al. 2010; Smith et al. 2013). To further elucidate the mechanisms of chemosensation through ASER, we characterized as yet uncloned che-5. We focused on che-5 because CB1073 che-5(e1073) mutant, the unique strain/allele of the gene, showed chemotaxis defects characteristic of ASER-specific malfunction; a severe defect in attraction to Cl–, whereas relatively moderate defect in Na+ chemotaxis (Lewis and Hodgkin 1977).
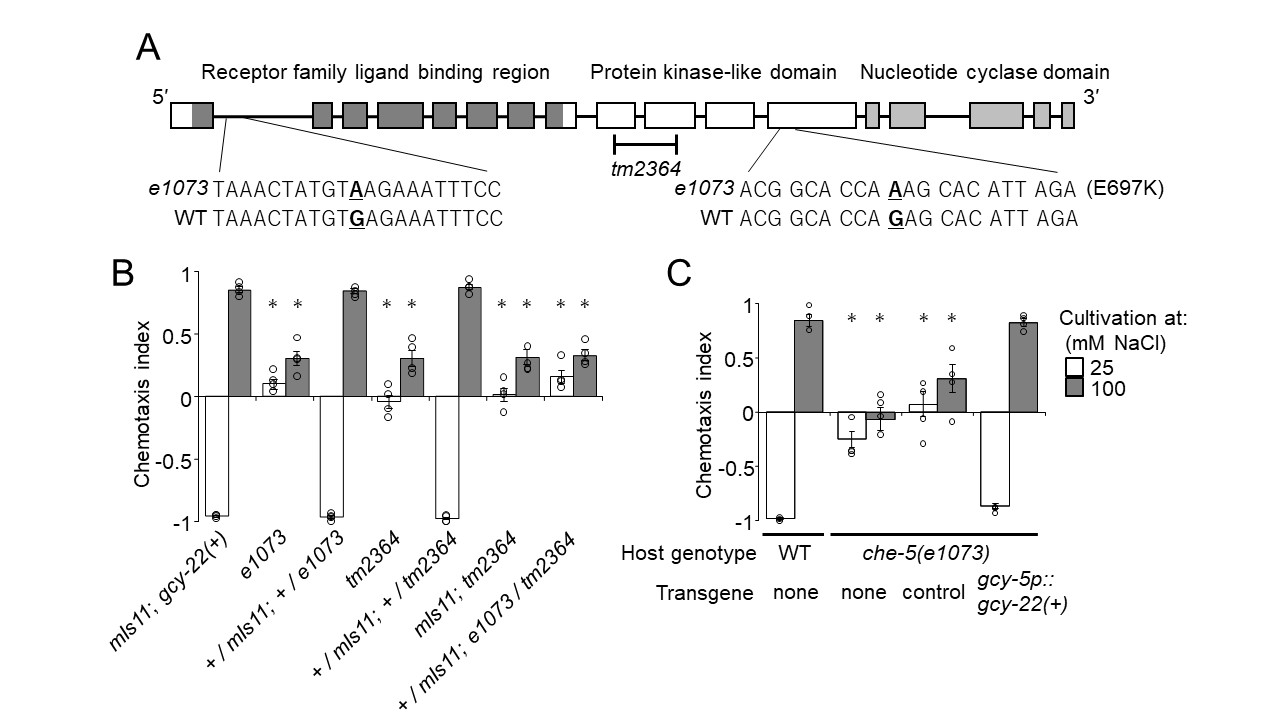
We mapped the mutation of che-5(e1073) responsible for salt chemotaxis defect between genetic positions 24.52 (single nucleotide polymorphism (SNP): WBVar00240760) and 25.54 (SNP: WBVar00053592) on chromosome V by using SNPs between N2 and CB4856 (Wicks et al. 2001). The mapped region contained an ASER-specific chemotaxis gene, gcy-22 (genetic position: 25.28). This result was unexpected from the initial report that mapped e1073 on chromosome IV (Lewis and Hodgkin1977), but consistent with a recent observation in which whole-genome sequencing failed to identify a mutation corresponding to che-5(e1073) on chromosome IV (Smith et al. 2013).
gcy-22(tm2364) harbors a deletion in the middle of gcy-22 coding region that results in a frame shift and therefore is a putative null allele (Fig. 1A). Salt concentration chemotaxis of the animals heterozygous for e1073 and tm2364 showed that the two alleles failed to complement each other, indicating that these alleles affect the same locus (Fig. 1B). Nucleotide sequencing of the gcy-22 locus revealed that e1073 carried ACG GCA CCA AAG CAC ATT AGA in which the adenine residue in bold letter was guanine in wild type (Fig. 1A). This transition results in a missense change E697K in GCY-22 isoform a. The glutamate residue is located in the kinase-like domain and well conserved in rGC proteins. In addition, CB1073 carried another nucleotide substitution within the first intron of gcy-22, TAAACTATGTAAGAAATTTCC, in which the adenine residue in bold letter was guanine in wild type (Fig. 1A). Furthermore, expression of a cDNA for gcy-22a in CB1073 in ASER-specific manner completely rescued the salt chemotaxis defect of the mutant (Fig. 1C). These results strongly indicate that che-5 is allelic to gcy-22 and the chemotaxis defect of e1073 is due to the mutations of gcy-22 locus.
Methods
Request a detailed protocolSalt concentration chemotaxis was evaluated as described (Kunitomo et al. 2013). A chemotaxis index was calculated to quantify the behavior as follows. Chemotaxis index = {(N at high NaCl-region) – (N at low NaCl-region)} / {(total N) – (N that did not move from the origin)}, in which N represents number of animals. For complementation tests, males of PD4792 (mIs11 with gcy-22(+) background) or JN2608 (mIs11 with gcy-22(tm2364) background) were mated with CB1073 hermaphrodites. F1 progenies were tested for salt concentration chemotaxis, and crossed progeny hermaphrodites that carried mIs11 were separately counted from self-progeny hermaphrodites that did not carry the marker. For rescue experiments, 5 ng/microL gcy-5p::gcy-22a construct was introduced into CB1073 with 15 ng/microL myo-3p::venus as a transformation marker.
Reagents
Strains. The JN strains are available upon request. Others are available at Caenorhabditis Genetic Center (CGC).
Bristol N2: wild type
CB4856: wild type
CB1073: che-5(e1073) V.
JN967: gcy-22(tm2364) V.
JN2606: che-5(e1073) V; peEx2606[myo-3p::venus].
JN2607: che-5(e1073) V; peEx2607[gcy-5p::gcy-22a myo-3p::venus].
JN2608: mIs11[myo-2p::GFP pes-10p::GFP gut-promoter::GFP] IV; gcy-22(tm2364) V.
PD4792: mIs11[myo-2p::GFP pes-10p::GFP gut-promoter::GFP] IV.
Acknowledgments
N2, CB4856, PD4792, and CB1073 were provided by the CGC. tm2364 was provided by the National Bioresource Project (NBRP)-Japan.
References
Funding
Japan Society for the Promotion of Science (JSPS) KAKENHI 19K06952 and The Salt Science Foundation No. 2043 to HK. JSPS KAKENHI JP17H06113, 19H04980 and Japan Science and Technology Agency (JST) CREST JP17H06113 to YI.
Reviewed By
Oliver HobertHistory
Received: September 19, 2020Revision received: September 23, 2020
Accepted: October 1, 2020
Published: October 8, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Kunitomo, H; Iino, Y (2020). Caenorhabditis elegans che-5 is allelic to gcy-22. microPublication Biology. 10.17912/micropub.biology.000313.Download: RIS BibTeX