Institute of Healthy Ageing and the Research Department of Genetics, Evolution, and Environment, University College London, London, U.K.
Pennsylvania State University, Center for Eukaryotic Gene Regulation, University Park, PA, U.S.A.
Clark H Smith Brain Tumour Centre, Arnie Charbonneau Cancer Institute, & Department of Biochemistry and Molecular Biology, University of Calgary, Alberta, Canada
Description
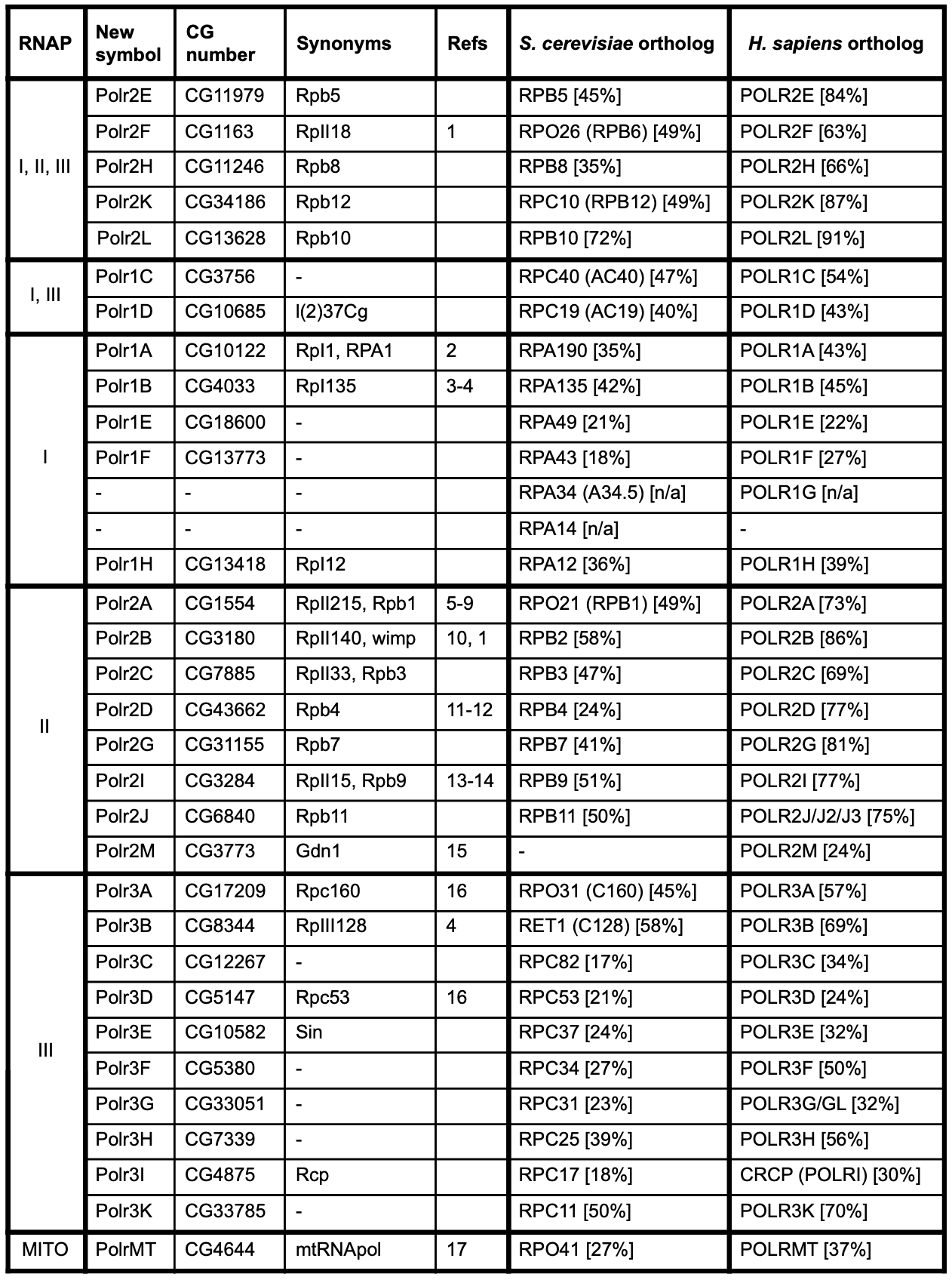
Three highly conserved, multisubunit RNA polymerase (RNAP) enzymes, RNAPs I, II, and III, transcribe the eukaryotic nuclear genome (reviewed by Cramer et al.. 2008, Vannini and Cramer 2012, Griesenbeck et al.. 2017, Cramer 2019). Each one synthesizes different classes of RNA from DNA templates: RNAP I synthesizes the ribosomal RNA precursor that is processed into most ribosomal RNAs (rRNAs), RNAP II makes messenger RNAs (mRNAs) and a variety of non-coding RNAs, and RNAP III synthesizes short, non-coding RNAs including transfer RNAs (tRNAs), the small 5S rRNA and the U6 small nuclear RNA. Each RNAP contains between 12–17 subunits, ten of which form a structurally conserved catalytic core with additional subunits located on the periphery. Notably, five subunits are shared among all three RNAPs and two others are shared between RNAPs I and III. In contrast to the nuclear RNAPs, a single subunit mitochondrial RNAP transcribes the rRNAs, mRNAs and tRNAs of the mitochondrial genome (Arnold et al.. 2012).
While much of what we know about eukaryotic RNAP composition and function comes from studies on yeast and human cells, several Drosophila melanogaster (hereafter, Drosophila) RNAP subunits have also been isolated and characterized, particularly by Greenleaf, Bautz and colleagues in the 1980s–90s (Greenleaf et al.. 1980, Searles et al.. 1982, Greenleaf 1983, Biggs et al.. 1985, Falkenburg et al.. 1987, Jokerst et al.. 1989, Kontermann et al.. 1989, Seifarth et al.. 1991, Hamilton et al.. 1993, Liu et al.. 1993, Knackmuss et al.. 1997). Since the publication of the Drosophila genome sequence in 2000, the genes encoding all the subunits of RNAP II (Aoyagi and Wassarman, 2000), several subunits of RNAPs I and III (see supplementary data of Filer et al.. 2017 and Martinez Corrales et al.. 2020) and the mitochondrial RNAP (Fernández-Moreno et al.. 2009) have been identified. Nevertheless, a systematic and complete survey of Drosophila RNAP genes is lacking, which has resulted in haphazard nomenclature within the fly literature and FlyBase (flybase.org, Thurmond et al.. 2019).
We employed a multi-pronged approach to systematically identify all genes encoding Drosophila RNAP subunits (see Methods for details). First, we obtained complete lists of RNAP subunits for yeast (Saccharomyces cerevisiae) and humans from recent publications and online resources, and used these to identify the Drosophila orthologs. Second, we obtained a list of all Drosophila genes annotated with relevant Gene Ontology (GO) terms. Importantly, these annotations include those based on direct experimental evidence as well as inferences based on sequence similarity/orthology and the presence of defined protein domains. Finally, we searched the Drosophila literature for reports of individual, or lists of, RNAP subunits. The results of these three approaches were cross-checked and integrated, and the results are presented in Table 1.
We find that a total of 31 distinct genes encode RNAP subunits in Drosophila. We identified genes encoding the five subunits shared between RNAPs I, II and III as well as the two subunits shared by RNAPs I and III. We also identified genes encoding an additional five subunits of RNAP I, an additional eight subunits of RNAP II, an additional ten subunits of RNAP III and the mitochondrial RNAP. Thus, Drosophila possesses twelve RNAP I subunits, thirteen RNAP II subunits, seventeen RNAP III subunits, and a single mitochondrial RNAP. Only a third of these have been characterized directly in Drosophila, either biochemically or genetically, with research having focussed on RNAP II subunits and the largest subunits of RNAPs I and III (see Refs column of Table 1). The Drosophila subunits show a range of 17–72% (mean of 39%) and 22–91% (mean of 55%) amino acid identity to their orthologs in S. cerevisiae and humans, respectively. A comparison of the complement of RNAP subunits across those three species reveals four notable differences: (i) Drosophila lacks an identifiable ortholog of yeast RPA34/human POLR1G (Martínez Corrales et al.. 2020); (ii) neither Drosophila or humans have an ortholog of yeast RPA14 (Russell and Zomerdijk 2006; Martínez Corrales et al.. 2020); (iii) yeast lack the POLR2M subunit, which defines a metazoan-specific RNAP II subpopulation (Hu et al.. 2006); and (iv) humans possess multiple copies of genes encoding RPB11/POLR2J and RPB7/POLR3G, whereas these are single-copy genes in Drosophila and yeast.
Prior to this study, 22 of the 31 Drosophila RNAP genes had been named in FlyBase using a variety of conventions. Seven were named based on the empirically determined molecular weight of the Drosophila proteins (RpII18, RpI1, RpI135, RpII215, RpII140, RpII15, RpIII128), following a nomenclature originally proposed in Greenleaf et al.. 1980. Fourteen RNAP genes had been named after their yeast or human ortholog, and one additional gene (Sin) was named for an unrelated physical interaction (Dong and Bell, 1999). The remaining nine genes were unnamed or had only a ‘placeholder’ symbol. We wished to assign an informative, systematic nomenclature to all Drosophila RNAP genes. Unfortunately, a universal eukaryotic RNAP nomenclature system does not exist, with two different systems currently in use for yeast and humans/vertebrates (Table 1). We propose that the human nomenclature system is adopted for the Drosophila genes in FlyBase for the following reasons: (i) individual Drosophila RNAP subunits show greater identity to the human subunits compared to yeast; (ii) the overall complement of Drosophila RNAP subunits is more similar to humans than yeast; (iii) unlike the yeast nomenclature, the human nomenclature follows a systematic format for all subunits; (iv) using the human nomenclature for the Drosophila subunits will facilitate the use/comparison of Drosophila data in biomedicine. (The yeast nomenclature will be retained/added to the Drosophila gene reports as searchable and browsable synonyms.)
In conclusion, our complete and rationalized listing of Drosophila RNAP subunits will be useful to Drosophila researchers working in this field as well as to those wishing to compare RNAP biology between fly, yeast, human and other species.
Methods
Request a detailed protocolPublications identifying/characterizing Drosophila RNAP subunits were identified using PubMed (pubmed.ncbi.nlm.nih.gov), FlyBase (flybase.org, Thurmond et al.. 2019) and Google (www.google.com). Published lists of S. cerevisiae and human RNAP subunits were obtained from Huang and Maraia 2001, Hu et al. 2002, Russell and Zomerdijk 2006, Cramer et al.. 2008, Vannini and Cramer 2012 and Griesenbeck et al.. 2017. In addition, a curated list of human RNAP subunits was obtained from the HGNC (www.genenames.org/data/genegroup/#!/group/726, Braschi et al.. 2019). Ortholog predictions and protein identity percentages were obtained from the integrative ortholog prediction tool, DIOPT (v8) (Hu et al.. 2011) via FlyBase. All reported orthologs in Table 1 are reciprocal best hits, with the exception of human POLR3G and POLR3G paralogs, where all genes are listed. Orthology predictions were verified using the HCOP tool (Eyre et al.. 2007). The Alliance of Genome Resources database (www.alliancegenome.org (release 3.1.1), The Alliance of Genome Resources Consortium, 2020) was used to query fly, yeast and human for relevant GO annotations (using terms RNA polymerase I complex (GO:0005736), RNA polymerase II, core complex (GO:0005665), RNA polymerase III complex (GO:0005666) and mitochondrial DNA-directed RNA polymerase complex (GO:0034245)). Gene symbol information was obtained from FlyBase (FB2020_03), SGD (www.yeastgenome.org, accessed 17th August 2020) and HGNC (www.genenames.org, accessed 17th August 2020).
Acknowledgments
We thank Kevin Cook, Julia Zeitlinger and Joan Conaway for comments on the manuscript, and Elspeth Bruford and Bryony Braschi at the HGNC for discussions on human RNAP gene nomenclature.
References
Funding
S.J.M. is funded by a grant from the National Human Genome Research Institute of the NIH [U41HG000739] to Norbert Perrimon (PI), Nicholas Brown (co-PI). N.A. is funded by grants from the BBSRC [BB/S014357/1 and BB/R014507/1], D.S.G. is funded by a grant from the National Institute of General Medical Sciences of the NIH [R01-GM0474777], and S.S.G is funded by a project grant from the Canadian Institutes of Health Research.
Reviewed By
AnonymousHistory
Received: September 21, 2020Revision received: October 12, 2020
Accepted: October 16, 2020
Published: October 20, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Marygold, SJ; Alic, N; Gilmour, DS; Grewal, SS (2020). In silico identification of Drosophila melanogaster genes encoding RNA polymerase subunits. microPublication Biology. 10.17912/micropub.biology.000320.Download: RIS BibTeX