Description
Caenorhabditis elegans feeds on bacteria in decomposing vegetation. Lipids, carbohydrates and proteins derived from microbes are digested into fatty acids, simple sugars and amino acids in C. elegans alimentary canal and absorbed by intestinal cells containing microvilli. Approximately 80% of fatty acids in C. elegans is derived from E. coli (Perez and Van Gilst, 2008). Nutrient limiting conditions can cause developmental delay in larvae (Cassada and Russell, 1975; Golden and Riddle, 1982) while complete starvation leads to L1 larval arrest or dauer formation (Baugh, 2013). Interestingly it has been reported that C. elegans fed on yeast Cryptococcus curvatus show developmental lag (Sanghvi et al., 2016) and growth arrest on Gram-positive bacterium Enterococcus faecalis (Garsin et al., 2001). We have recently shown that E. faecalis infection causes lipid droplet utilization in adult C. elegans, a process termed immunometabolism (Dasgupta et al., 2020). In this study, we have investigated the developmental arrest induced by E. faecalis in C. elegans larvae to show that the arrest is induced at L1 and L2 larva stage.
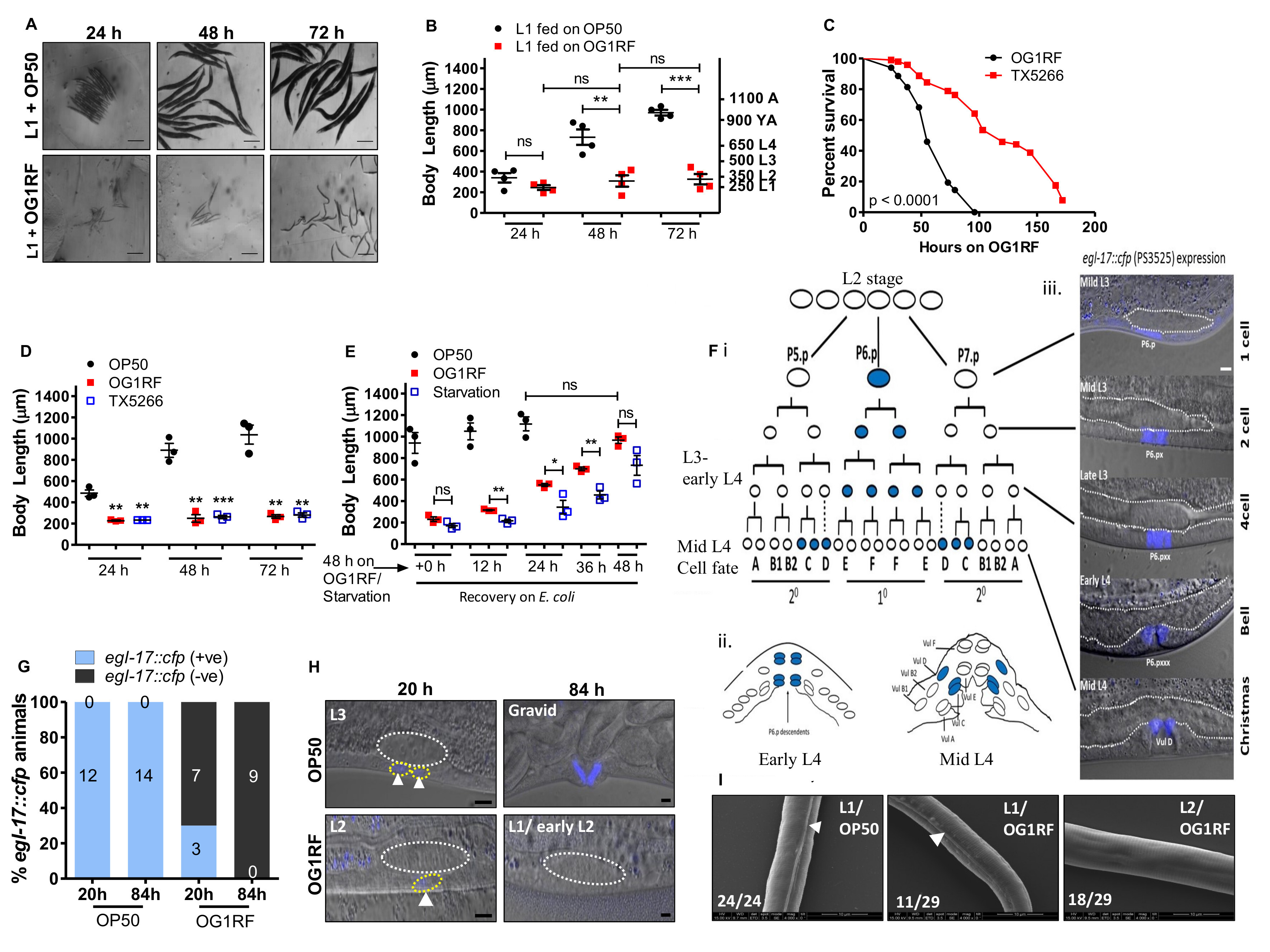
To determine the developmental stage at which larvae get arrested upon feeding on E. faecalis OG1RF, we exposed a synchronized population of L1 larvae (250 μm body length) to either E. coli OP50 or E. faecalis OG1RF diet and measured the body length at 24, 48 and 72 hours of feeding on each bacterium (Figure 1A and 1B). We observed that C. elegans fed on OG1RF diet were shorter than animals fed on OP50 at all time points (Figure 1B). At 72 hours of feeding, OP50-fed animals had reached adult body lengths measuring >1000 μm. However, OG1RF-fed animals had body length ranging between 250-350 μm, equivalent to L1 and L2 larval stages, indicating an early developmental arrest. Next, we set out to ask if larval arrest on E. faecalis diet was due to virulence of E. faecalis. The Fsr locus in E. faecalis encodes a two-component system (response regulator FsrA, peptide lactone FsrB and histidine kinase FsrC) to control production of virulence factors, gelatinase and serine protease (Qin et al., 2000). ΔfsrB E. faecalis (TX5266 strain) is attenuated for virulence in C. elegans (Figure 1C) as shown earlier (Garsin et al., 2001; Qin et al., 2000). To test if E. faecalis virulence was a cause for arrest, we fed L1 larvae with TX5266 strain and found that it caused larval arrest similar to the arrest caused by OG1RF feeding (Figure 1D). This indicated that factors other than pathogenesis of E. faecalis are likely responsible for developmental arrest in C. elegans. To understand if E. faecalis-induced larval arrest was reversible, we allowed 48 h starved larvae, 48 h OG1RF-fed larvae and 48 h OP50-fed larvae to resume/continue feeding on OP50 for additional 12, 24, 36 and 48 hours (Figure 1E). As shown, both starved and E. faecalis arrested larvae resumed larval development when fed E. coli diet, although starved larvae were slower in resuming development.
To further confirm that E. faecalis induced arrest during larval development, we also studied proliferation and lineage progression in hermaphrodite vulva (Figure 1F-1H). Development of vulva is tightly synchronized with larval development. In C. elegans hermaphrodite, vulva development from vulval precursor cells (VPC) is dependent on signalling from the gonadal cell called the Anchor Cell (AC). AC secretes LIN-3/EGF (Epidermal Growth Factor) to induce vulva fate in VPCs- P5.p, P6.p and P7.p- of the equipotent P(3-6).p lineage cells. P6.p, the VPC closest to anchor cell adopts 10 cell fate, and VPCs P5.p/P7.p adopt 20 fates. The EGL-17/FGF (Fibroblast Growth Factor) is induced transcriptionally by inductive and notch signalling in vulva cells (Burdine et al., 1998; Sulston and Horvitz, 1977). From early L3 to late L3, EGL-17 is expressed in P6.p/10 lineages and at mid L4 it switches to 20 lineage cells, VulC and VulD (Figure 1F). We followed P6p fate in OP50 and OG1RF-fed larvae. At 20 hour post-feeding (time prior to VPC P6.p division), 100% of OP50-fed larvae showed positive egl-17::cfp expression corresponding to L3 larval stage. However, in OG1RF-fed larvae only 30% larvae showed weak expression in P6.p cell corresponding to late L2 stage while 70% had no GFP expression indicating an L1 or early L2 stage. At 84 hours of feeding, we observed egl-17::cfp expression in vulD corresponding to adult stage in OP50-fed animals, but we observed no cell division in P6.p lineage in OG1RF–fed larvae indicating arrest at L1 or early L2 stages (Figure 1G and 1H). These observations confirmed that the vulva development in OG1RF-fed larvae was indeed arrested at early larval stages as VPCs did not divide and never acquired vulva fates. For the final confirmation, we also examined OG1RF arrested larvae for the presence of alae, longitudinal ridges in the cuticle which are observed only in L1, dauer and adult stage of C. elegans (Cox et al., 1981). We found that 72h-fed OG1RF larvae comprised of a mixed population of L1s and L2s, with 38% (11 out of 29) animals showing alae (L1) and the remaining 62% (18 out of 29) did not have alae (L2) (Figure 1I).
Taken together, our study shows that E. faecalis OG1RF cocci diet poorly supports growth of C. elegans larvae and cause development arrest at early larval stages.
Methods
Request a detailed protocolNematode maintenance
C. elegans were maintained at 20°C on nematode growth medium (NGM) seeded with OP50 E. coli. Wild type N2 was obtained from Caenorhabditis Genetics Center, Minnesota, USA. Strains used in this study have been listed in Table 2 (mentioned in the reagents section). All feeding assays were conducted at 25˚C.
Bacterial strains and Growth media
Bacterial strains used in this study are listed in Table 2. E. coli OP50 was grown in LB at 37˚C for 8 hours and seeded on NGM plates containing streptomycin. E. faecalis strains were grown in BHI broth with appropriate antibiotics for 5 hours at 37˚C and 50 μl culture was spread on BHI agar plates with respective antibiotics and kept at 37˚C overnight. Gentamycin (50 μg/ml) was used for E. faecalis OG1RF and Rifampicin (100 μg/ml) was used for TX5266 strain (Table 2).
L1 synchronization
NGM agar petri plates with 150 gravid adults each, were prepared for bleaching (2-3 petri plates for body length measurement and 10-12 petri plates for SEM experiment). Animals were washed with phosphate buffer saline (PBS) in a 15ml tube, after which a 1:1 mixture of 2X Bleach solution and PBS was added. Bleach solution was prepared by adding 900 μl sodium hypochlorite solution, 700 μl distilled water and 400 μl of 5N NaOH. The tube was shaken vigorously for 1-2 minutes until eggs were released into the solution. The eggs were immediately washed with 10 ml PBS and collected by centrifugation at 3000 rpm for 2 minutes. This step was repeated 5 times, following which eggs were resuspended in 5 ml PBS and maintained on a rotator at room temperature, for 20 hours for eggs to hatch to L1 larvae. L1s were spotted on E. coli and E. faecalis lawns, for carrying out further experiments.
Body length measurements
Synchronized L1 larvae (Figure 1A, B and D) were spotted on E. coli and E. faecalis lawns. Every 24 hours, 15-20 animals/time point/diet were mounted on 2% agarose pad and imaged using AxioCam I Cm1, Zeiss microscope. Body length of animals was measured using Image J software. Each experiment was repeated at least three times. For larval rescue experiment, L1s were spotted on E. coli, E. faecalis lawns or on plate NGM agar plates (for starvation) for 48 hours, after which they were transferred back to E. coli lawn and imaged after every 12 hours.
Vulva Development study
PS3525 strain (egl-17::cfp + unc-119(+)) was used to trace vulval precursor cell lineage during development. Synchronized L1s of PS3525 strain were exposed to E. coli and E. faecalis and imaged at 20 h and 84 h post exposure using LSM880 airyscan microscope. The number of vulva cells showing egl-17::cfp fluorescence were counted. 10-20 animals were used for each time point for each diet. Vulval precursor cell lineage is described (Figure 1, F-H.).
C. elegans survival assay
To assess the survival of C. elegans during infection, we performed survival assays. A single E. faecalis bacterial colony was inoculated in 2 ml of BHI broth. 50 μl of bacterial culture was spread on 60 mm BHI agar plates with appropriate antibiotic and incubated at 37˚C overnight. 100-120 synchronized young adult animals were exposed to E. faecalis at 25°C and scored for survival at the times indicated in Figure 1C. Animals were considered dead when they failed to respond to touch. Each survival assay was performed three times.
Scanning Electron Microscopy
Synchronized L1s exposed to E. faecalis lawn for 72 hours and E. coli lawn for 3-4 hours, were washed with phosphate buffer saline (PBS) three times and collected. These animals were incubated in fixation buffer (5% glutaraldehyde, 5% formaldehyde and 0.2M HEPES, pH 7.3) overnight at room temperature. Next day animals were postfixed in a solution containing 2% osmium tetroxide, 200mM CaCl2 and 12.5% K3[Fe(CN)6] for 4 hours at room temperature. Animals were washed with sodium phosphate buffer three times, followed by four washes with water. Next, the animals were dehydrated through increasing concentration of ethanol and finally suspended in 100% ethanol overnight. This protocol was modified from (Shemer et al., 2004). The following day, animals were spotted on a coverslip, coated with gold and imaged using FEI XL-30 ESEM scanning electron microscope at Indian Institute of Science Advanced Microscopy Facility.
Statistics
Statistical analysis was done using Graphpad Prism software. Unpaired t-test was used for comparison of mean body lengths obtained from measurements across individual experiments (N) on populations composed of 15-20 individuals (n). Kaplan-Meier analysis was performed to calculate survival fractions and the Log Rank test to compare survival curves (see Table 1). TD50, the time to death for 50% of the population was calculated as described (Singh and Aballay, 2006). Survival curves were considered different from the appropriate control when P values were < 0.05.
Reagents
Table 1. Statistical analysis of Kaplan Meier Survival Curves.
| Pathogen Used | Trial no | Strain Genotype | TD50
(Hours) |
No. of animals used / No. of animals censored | Statistical significance |
| E. faecalis | I | WT OG1RF
WT ΔfsrB OG1RF |
54
110 |
100/32
104/40 |
P < 0.0001 |
| E. faecalis | II | WT OG1RF
WT ΔfsrB OG1RF |
51
74 |
100/36
100/49 |
P < 0.0001 |
| E. faecalis | III | WT OG1RF
WT ΔfsrB OG1RF |
52
86 |
100/24
100/21 |
P < 0.0001 |
| E. faecalis | IV | WT OG1RF
WT ΔfsrB OG1RF |
44
98 |
100/41
100/55 |
P < 0.0001 |
Table 2. List of strains used in this study.
| S. no. | Strain name | Source |
| 1. | Caenorhabditis elegans N2 (Bristol) | Caenorhabditis Genetics Center (CGC), University of Minnesota |
| 2. | C. elegans PS3525 egl-17::cfp + unc-119(+) | Caenorhabditis Genetics Center (CGC), University of Minnesota |
| 3. | E. coli OP50. Uracil auxotroph. E. coli B | A gift from Dr. Alejandro Aballay, Oregon Health and Science University (OSHU) |
| 4. | E. faecalis OG1RF | A gift from Dr. Alejandro Aballay, OSHU |
| 5. | E. faecalis TX5266 ΔfsrB OG1RF | A gift from Dr. Barbara Murray, McGovern Medical School, UT Health |
Acknowledgments
We thank Dr. Barbara E. Murray and Dr. Alejandro Aballay for Enterococcus faecalis strains. All C. elegans strains were provided by CGC which is funded by the NIH Office of Infrastructure Programs (P40 OD01440).
References
Funding
The Wellcome Trust DBT India Alliance (Grant no. IA/I/13/1/500919)
Reviewed By
Ryan BaughHistory
Received: September 25, 2020Revision received: October 13, 2020
Accepted: October 16, 2020
Published: October 27, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Dasgupta, M; Bojanala, N; Shashikanth, M; Singh, V (2020). Caenorhabditis elegans larvae undergo early developmental arrest on a diet of Gram-positive bacterium Enterococcus faecalis. microPublication Biology. 10.17912/micropub.biology.000321.Download: RIS BibTeX